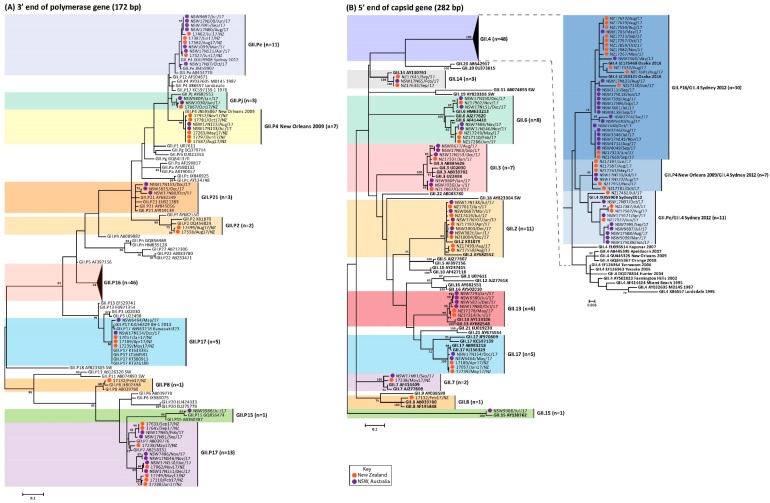
Figure 4.
Phylogenetic analysis of polymerase (RdRp) and capsid (VP1) regions of GII norovirus. Representative norovirus GII strains isolated in this study (n = 92) from both NSW, Australia and New Zealand, are shown in this phylogenetic analysis. They are denoted with a colored bullet (•), where Australian samples are represented in purple and New Zealand samples in orange. All samples are labelled with their geographical location and time of strain isolation. Due to the number of sequences grouped within GII.P16 and GII.4, the sequences within those clusters were compressed and represented by the black triangles. Reference strains were obtained from the GenBank database, labelled with their genotype and accession number. (A) Maximum likelihood phylogeny derived from partial 3′ end of polymerase gene (172 bp) of GII noroviruses. (B). Maximum phylogeny derived from partial 5′ end of capsid gene (282 bp) of GII noroviruses. Sequence alignments were performed with MUSCLE algorithm. Maximum likelihood phylogenetic trees were produced using MEGA 5 software with bootstrapping test of 1000 replicates, based on the Kimura 2-parameter model. The bootstrap percentage values are shown at each branch point for values ≥70%. The number of substitutions per site is indicated by the scale bar.