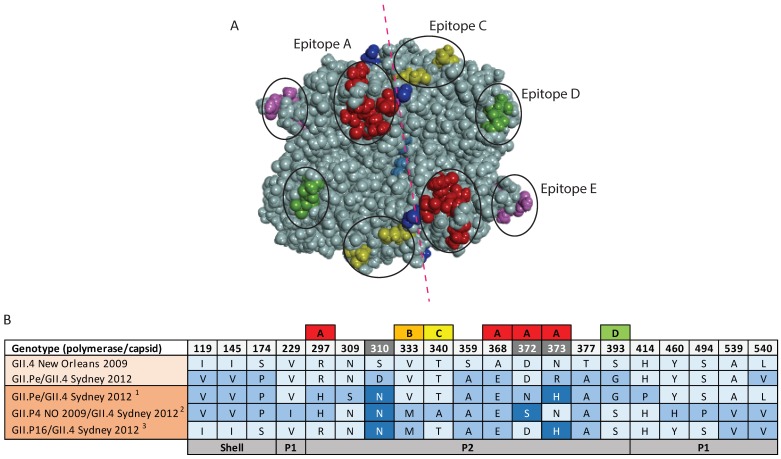
Figure 6.
Capsid residue variation and antigenic variation within the full-length capsid of GII.4 recombinant viruses. Full-length capsid sequence of novel GII.4 recombinant viruses were collected between July 2014 and December 2017. Consensus sequences of contemporary GII.Pe/GII.4 Sydney 20121 (n = 20), GII.4 New Orleans 2009/GII.4 Sydney 20122 (n = 13) and GII.P16/GII.4 Sydney 20123 (n = 9) were generated and compared to pandemic variants (New Orleans 2009 and Sydney 2012) for the identification of antigenic variations, especially within the P2 protruding domain. (A) Characterized blockade antibody epitopes are highlighted in the different colors; epitope A (red), epitope C (yellow), epitope D (green) and epitope E (purple). The residues within P2 domain, especially those within epitope A, which differed from the pandemic Sydney 2012 variant. The red dotted line shows symmetry. (B) Antigenic variations observed between full length capsid sequences of pandemic Sydney variant and the new Sydney 2012 recombinants. Labelled boxes above the antigenic sites indicate sites within known blockade epitopes A–D that are important determinants of viral antigenicity. The A–D epitopes identified in GII.4 capsid are colored; epitope A (red), epitope B (orange), epitope C (yellow) and epitope D (green). The numbers across the top panel indicate the amino acid position within the VP1 sequence, hypervariable sites with ≥3 amino acids substitutions across all sequences are shaded in grey. Residues that vary from the GII.4 New Orleans 2009 sequence over time are indicated by shades of blue. The bottom panel indicates the positions of the shell, P1 and P2 domains within the VP1 capsid protein. The prototype of each pandemic/recombinant variants are indicated in the lightest shade of orange.