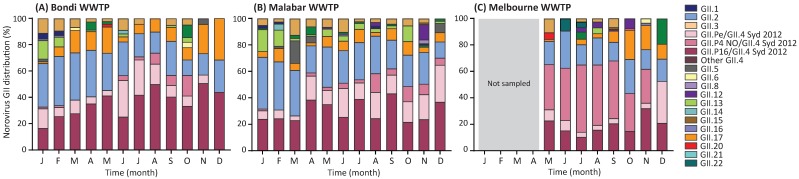
Figure 7.
Norovirus GII genotype distribution in wastewater samples collected from Sydney and Melbourne, 2017. The norovirus GII capsid genotypic distribution was determined in wastewater samples by amplicon sequencing using NGS technology. After amplification of the capsid region, a second round PCR was performed for the addition of adapters prior NGS library preparation. Libraries were sequenced on the MiSeq platform and an average of 152,879 reads were generated for each sample. Geneious was used for merging and mapping of the reads to the norovirus GII reference sequences. (A) The monthly genotype distribution of norovirus GII viruses in wastewater samples collected from Bondi WWTP in Sydney, Australia. (B) Norovirus GII genotypic diversity was examined in wastewater samples collected monthly from Malabar WWTP in Sydney, Australia. (C) The monthly norovirus GII genotype diversity was also examined in Melbourne, all samples were collected from western wastewater treatment plant. Samples were not collected between January and April 2017. Norovirus GII capsid genotype and GII.4 recombinants are labelled in different colors as indicated by the legend.