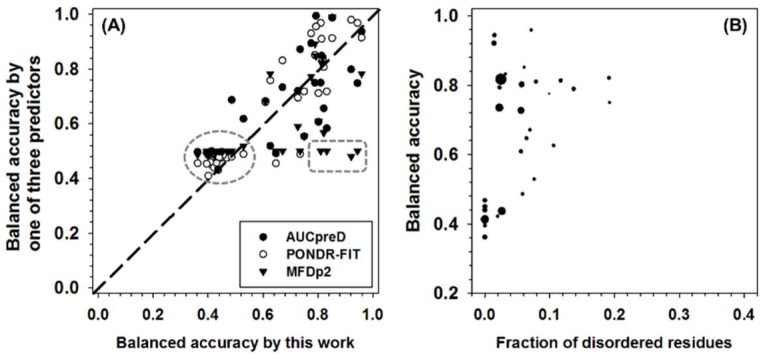
Figure 5.
(A) Comparison of per-sequence balanced accuracy among AUCpreD (filled circle), PONDR-FIT (open circle), MFDp2 (filled triangle), and this work on sequences in the CASP10 test dataset. The reasons for selecting these predictors are: (1) they are developed in recent years; (2) they have higher performance on some of the accuracy measures; (3) for simplicity of visualization, only four predictors were selected. The x-axis shows the per-sequence balanced accuracy of this work, and the y-axis shows the per-sequence accuracy of the other three predictors. (B) Per-sequence balanced accuracy of this work (y-axis) as a function of the fraction of experimentally validated intrinsically disordered amino acids (IDAAs) (x-axis). The size of the symbol is proportional to the length of the sequence.