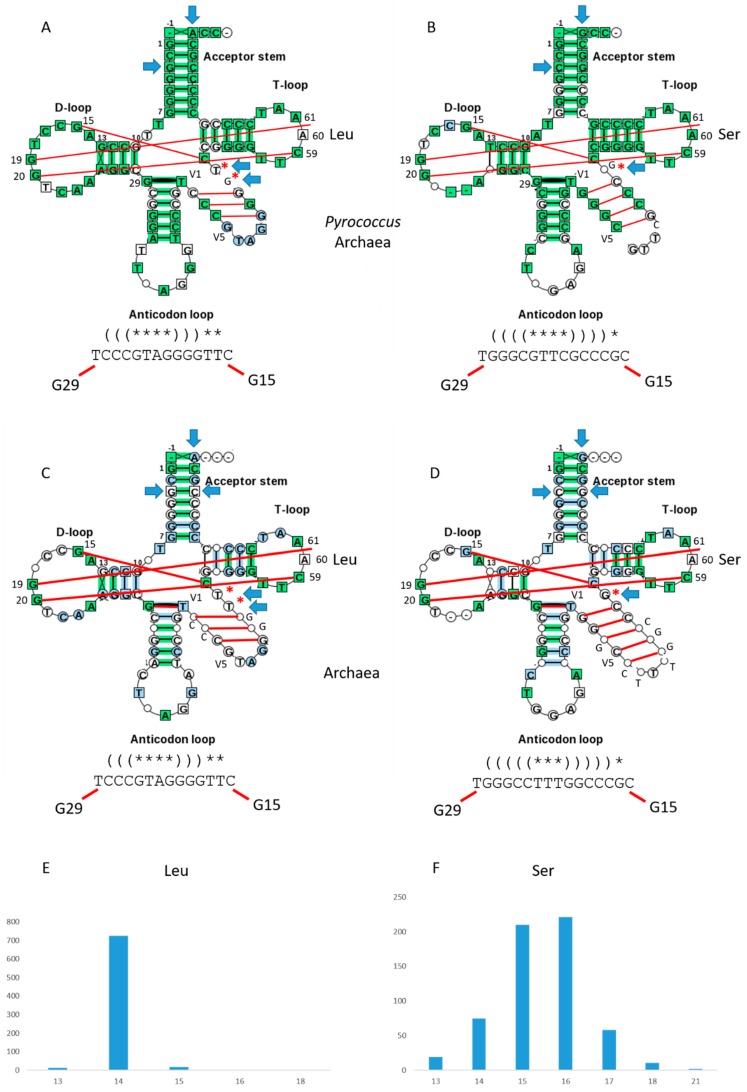
Figure 4.
Typical type-II tRNAs in archaea. (A) tRNALeu in Pyrococcus. (B) tRNASer in Pyrococcus. (C) tRNALeu in archaea. (D) tRNASer in archaea. Some interactions within the D loop, V loop and T loop are indicated with red lines. Blue arrows indicate determinants (or anti-determinants) for discrimination of tRNAs by aaRS enzymes. Red asterisks indicate V loop bases not in the V loop stem, that may allow discrimination of different V loops (i.e., by LeuRS, SerRS and other aaRS enzymes). Note that Pyrococcus tRNASer has two perfect UAGCC repeats in the D loop (8-UAGCCUAGCC-17). (E) Histogram of N for tRNALeu in archaea. (F) Histogram of N for tRNASer in archaea.