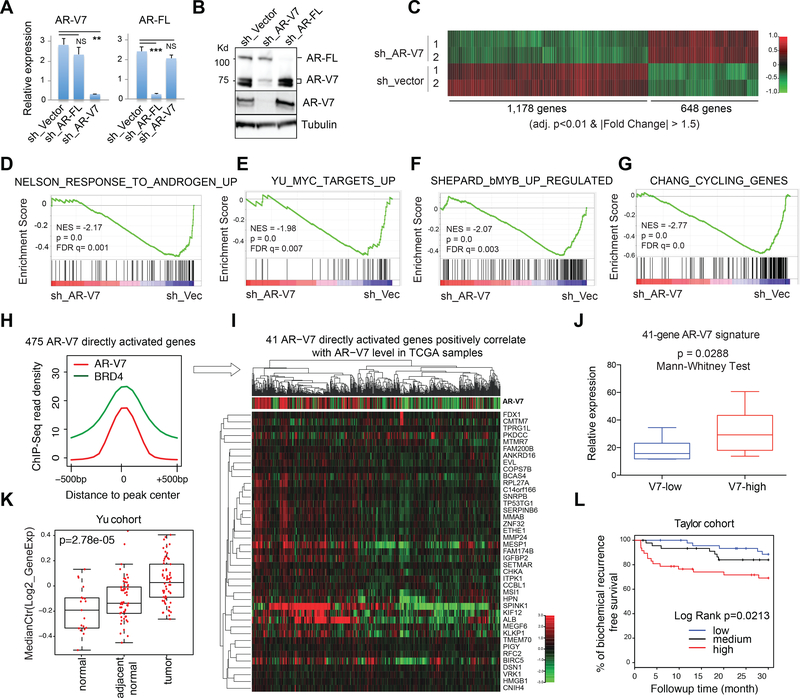
Fig 2. Integration of genomic datasets from 22Rv1 CRPC cell model and clinical prostate cancer samples reveals the AR-V7 direct or unique target signature predicting prognosis.
(A-B) RT-qPCR (A) and immunblot (B) show selective KD of AR-FL or AR-V7 in 22Rv1 cells. Used in B are AR N-terminus (top; pan-AR) and AR-V7-specific (middle) antibodies. NS, not significant, **, p<0.01; ***, p<0.001.
(C) Heatmap showing expression of genes down (left) and up regulated (right) after AR- V7 KD (sh_V7) relative to vector in ligand-starved 22Rv1 cells (2 biological replicates per group). Threshold of differential expression is adjusted DESeq p value (adj. p) of <0.01 and fold-change (FC) of >1.5. Color bar, log2(FC).
(D-G) GSEA shows negative correlation of the indicated gene set with selective KD of AR-V7, relative to mock (sh_Vec).
(H) Averaged AR-V7 and BRD4 ChIP-Seq signals at the 475 genes that are upregulated by AR-V7 and also have direct AR-V7 binding in 22Rv1 cells.
(I) Heatmap showing that the 41 genes directly upregulated by AR-V7 in 22Rv1 cells also positively correlates (spearman rank r >0.2 and BH FDR <0.01) with AR-V7 expression level in the TCGA prostate cancer cohort. Top of panel I shows the ratio of RNA-Seq read counts specific to AR-V7 (i.e. those of CE in Fig 1A) to those common to all AR isoforms (i.e. AR N-terminal domain).
(J-K) Box plots shows mean expression values of the 41-gene AR-V7 direct targets (41- gene signature defined in I) in patient cohorts reported in (Beltran et al., 2016) (J) or (Yu et al., 2004) (K). V7-low/high, bottom/top one-third of patients ranked by AR-V7 expression in the cohort. The p-value and test are denoted on top.
(L) Kaplan-Meier curve for the above defined 41-gene AR-V7 direct target signature in a patient cohort reported in (Taylor et al., 2010).