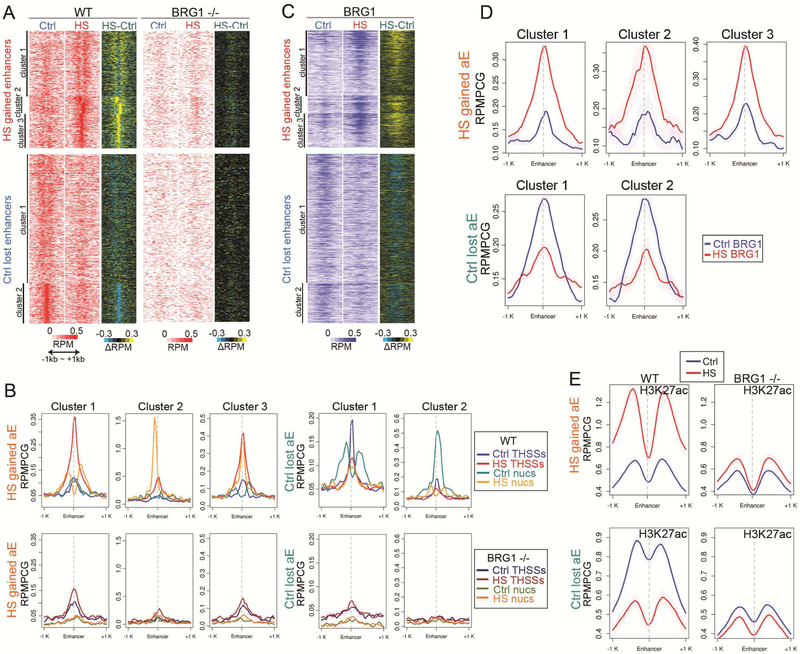
Figure 2. Alterations in enhancer activity induced by temperature stress involve nucleosome remodeling by BRG1.
(A) Repositioning of nucleosomes flanking HS gained enhancers observed in control H9 cells after temperature stress, based on ATAC-seq nucleosomal (180~247bp) read signal at enhancer summits ±1 kb (top). BRG1−/− cells fail to remodel nucleosomes at the same enhancers in after heat shock. Enhancers that become inactive after temperature stress lose positioned nucleosomes in control H9 cells based on ATAC-seq nucleosomal read signal at enhancer summits ±1 kb (bottom). BRG1−/− cells lack well positioned nucleosomes at enhancers lost after temperature stress even in control cells. Subtraction heatmaps included for comparison. RPMPCG: reads per million per covered region in 50 bp bin.
(B) Average profiles of ATAC-seq THSSs and nucleosomes on HS gained and lost enhancers in H9 cells in Ctrl and HS (top) show repositioning of nucleosomes, which is impaired in BRG1−/−H9 cells. Clusters derived by K-means clustering of nucleosomal reads. RPMPCG: reads per million per covered region in 50 bp bin
(C) Distribution of BRG1 ChIP-seq signal at enhancers gained (top) or lost (bottom) after temperature stress; anchors are in the same order as (D). Subtraction heatmaps included. RPMPCG: reads per million per covered region in 50 bp bin
(D) Average profiles of BRG1 ChIP-seq signals on the same anchors as (B). RPMPCG: reads per million per covered region in 50 bp bin
(E) Distribution of H3K27ac ChIP-seq signal at enhancers gained (top) or lost (bottom) in control and temperature stressed wild type and BRG1 KO cells; anchors are in the same order as (B). RPMPCG: reads per million per covered region in 50 bp bin.