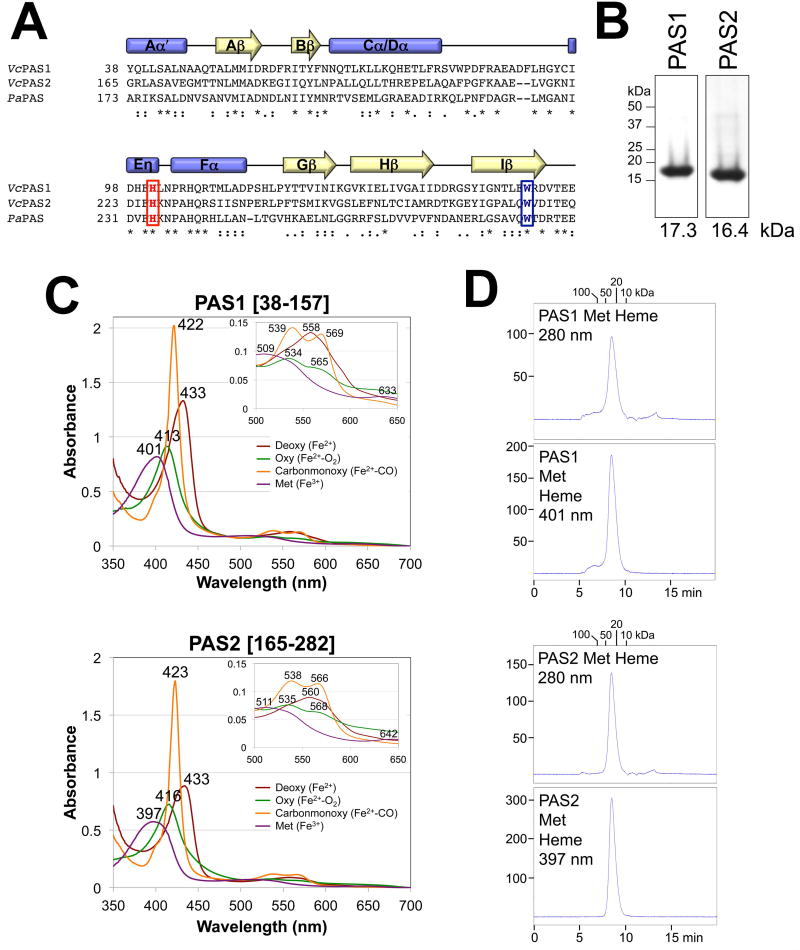
Fig. 3. Secondary structure, heme binding and oligomeric state of the PAS1 and PAS2 domains of VcAer2.
A. Sequence alignment of PAS1 and PAS2 from V. cholerae Aer2 and PaPAS from P. aeruginosa Aer2 as generated in ClustalW. The conserved His that coordinates heme in PaPAS, and the conserved Trp that stabilizes O2-binding to PaPAS, are highlighted red and blue, respectively. Secondary structure elements are based on the solved structures of PaPAS (Sawai et al., 2012, Airola et al., 2013a). Stars indicate conserved residues, colons indicate similar amino acids, and periods indicate amino acids with weakly similar properties.
B. Coomassie-stained SDS-PAGE of 5 µg of purified PAS1 [38–157] and PAS2 [165–282] peptides.
C. Absorption spectra of 10 µM purified PAS1 and PAS2 domains in the reduced (deoxy), oxidized (met), carbon monoxide-bound (carbonmonoxy) and oxygen-bound (oxy) states. The wavelength for each absorbance maximum is indicated. The insert shows an expanded view of peaks between 500 and 650 nm.
D. Elution profiles of isolated PAS1 and PAS2 peptides (200 µg) in their met-heme states during size-exclusion chromatography. Elution profiles are shown in arbitrary units at 280 nm to reveal total protein content (top panels) and at 401 nm (PAS1) or 397 nm (PAS2) to detect the elution of met-heme (bottom panels). The area under the peak for PAS2 is 97% the area of PAS1. Fractions were removed and analyzed by Western blotting; in all cases, PAS peptide co-eluted with the heme (not shown).