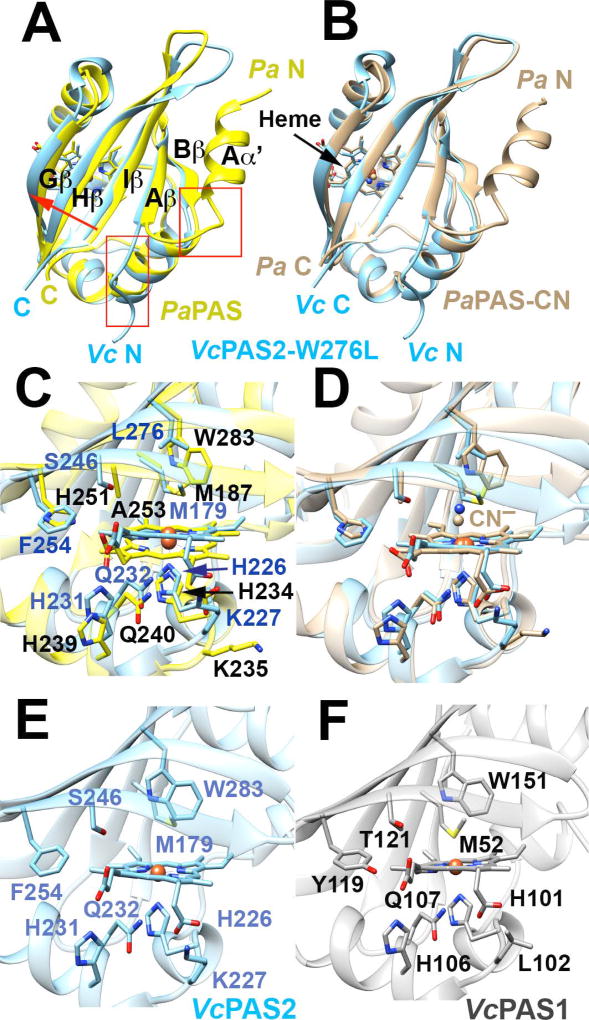
Fig. 7. The structure of PAS2-W276L and its relationship to PaPAS structures and WT PAS1 and PAS2 homology models.
A–B. Superposition of PAS2-W276L (A; PDB code: 6CEQ, blue) and PaPAS in the unliganded ferric-heme form (A; PDB code: 4HI4, yellow) and the CN−-bound form (B; PDB code: 3VOL, brown). The structures of PAS2-W276L and both PaPAS structures are similar, except that the Aα′ helix is dissociated and unstructured in PAS2-W276L (red boxes). However, PAS2-W276L is most similar to the ligand bound form of PaPAS, with the β-sheet shifted slightly relative to the ferric form of PaPAS (A; red arrow).
C–D. Superposition of the heme-binding pockets of PAS2-W276L (blue) and PaPAS in the unliganded ferric-heme form (C; yellow) and CN−-bound form (D; brown).
E–F. Comparison of the heme pockets of WT PAS2 (E) and WT PAS1 (F) homology models. Modeling of the WT PAS2 structure and PAS1 domain was accomplished in SWISS-MODEL using the PAS2-W276L structure as a template in both cases.