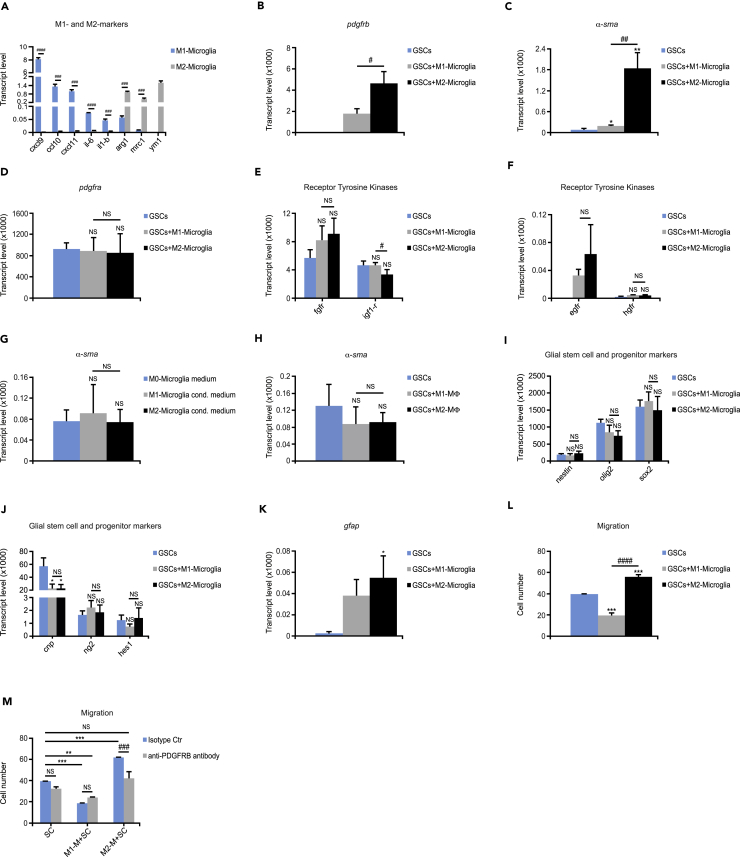
Figure 6.
TAMMs Induce Mouse Glioma Cells to Express PDGFRB
(A) Microglia were polarized to either an M1 or M2 phenotype. Graph shows gene expression levels of cxcl9, cxcl10, cxcl11, il-6, il-1b, arginase1, mrc1, and ym1 analyzed by qRT-PCR (n = 3).
(B–F) GFP+GSCs were co-cultured with M1- or M2-polarized microglia. Graphs depict qRT-PCR analysis of gene expression levels of flow-sorted tumor cells for (B) pdgfrb, (C) α-sma, (D) pdgfra, (E) fgfr and igf-1r, and (F) egfr and hgfr (n = 3).
(G) GFP+GSCs were cultured with M0 (not polarized)-, M1- or M2-like microglia conditioned media. Graph displays gene expression levels for α-sma quantified by quantitative RT-PCR (n = 3).
(H) GFP+GSCs were co-cultured with M1- or M2-polarized BMDMs. Graph shows qRT-PCR analysis of gene expression levels of flow-sorted tumor cells for α-sma (n = 3).
(I–K) GFP+GSCs were co-cultured with M1- or M2-polarized microglia. Graphs depict quantitative RT-PCR analysis of gene expression levels of flow-sorted tumor cells for (I) nestin, olig2, and sox2; (J) cnp, ng2, and hes1; and (K) gfap (n = 3).
(L and M) GFP+GSCs were co-cultured with M1- or M2-polarized microglia, and migrated tumor cells were stained for the OLIG2 marker. Graphs display (L) migrated tumor cells (n = 3) and (M) migrated tumor cells with isotype control or neutralizing PDGFRB antibody (n = 3).
Statistical analysis: Student's t test (A) and one-way ANOVA (B–M) were used: *p < 0.05, **p < 0.01, ***p < 0.001; * indicates significance (B–L) compared with tumor cells alone, and (M) compared with tumor cells without PDGFRB blockage; #p < 0.05, ##p < 0.01, ###p < 0.001, ####p < 0.0001; # indicates significance between (A) M1-like and M2-like microglia and (B–L) tumor cells co-cultured with M1 microglia compared with M2 microglia and (M) tumor cells co-cultured with M2-like microglia without PDGFRB blockage compared with PDGFRB blockage. All data represent one out of three independent experiments and are presented as the mean +SD. See also Table S1.