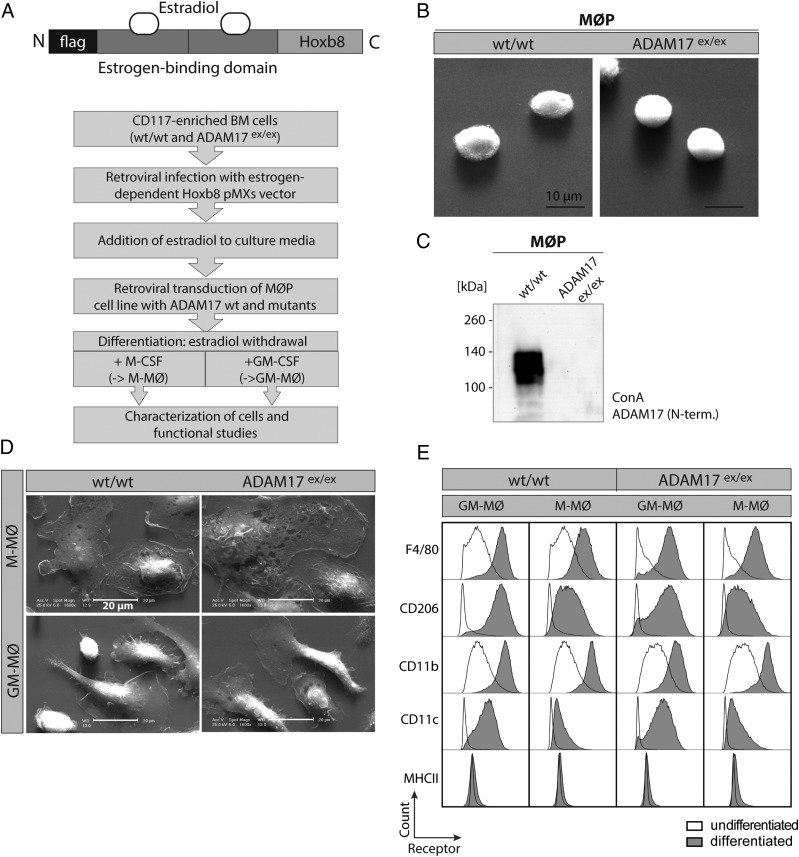
FIGURE 2.
Establishment and characterization of MØP on wt and ADAM17ex/ex background. (A) Flow diagram of protocol to establish MØP from wt and ADAM17ex/ex mice and further differentiation to M-MØ and GM-MØ by application of M-CSF and GM-CSF. Top, Scheme of the retroviral HoxB8 expression plasmid, which is used for estrogen-dependent immortalization of CD117-positive bone marrow–derived cells. (B) Electron microscopic picture of HoxB8-immortalized macrophage progenitor cells gained from wt and ADAM17ex/ex mice, indicating equal shape and size of MØP. Scale bar, 10 μm. (C) Representative immunoblot of Con A–enriched ADAM17, validating protein expression in wt but not in ADAM17ex/ex MØP using an N-terminal (10.1) Ab. (D) EM was performed on differentiated M-MØ and GM-MØ from wt and ADAM17ex/ex MØP. M-MØ from either genetic background exhibit characteristic round cell structure, whereas the GM-MØ show a narrower cell shape with extensions. Scale bar, 20 μm. (E) FACS analysis of cell surface markers of differentiated M-MØ and GM-MØ in wt and ADAM17ex/ex background in comparison with undifferentiated MØP. Shaded histograms represent the receptor staining of differentiated M-MØ or GM-MØ. Bold lines indicate receptor signal of undifferentiated MØP. Signal of the receptor staining increases according to differentiation and is independent of ADAM17 presence. The histograms are representative of plots from three independent experiments for each marker.