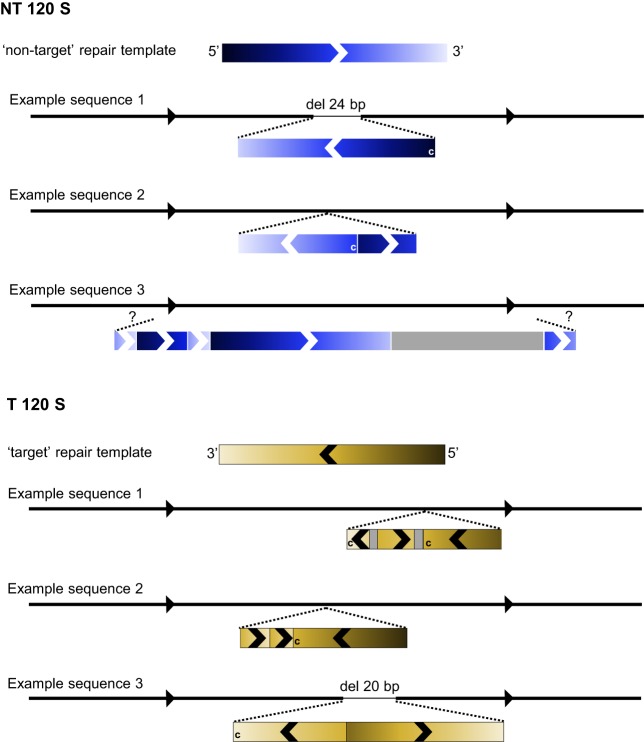
Fig. 2.
Simplified schematic representation of erroneous repair-template integration events at the smad6a zebrafish gene. For both the ‘NT 120 S’ and ‘T 120 S’ repair templates, three examples of NGS reads are schematized to clarify the erroneous repair patterns that were encountered in this study. For each example, the black line with arrows represents the reference sequence, and the two black dashed lines indicate the approximate location of repair-template insertions. Inserted fragments are depicted as rectangles, colored in blue or yellow, in accordance with the used repair template (indicated in the scheme as ‘non-target’ or ‘target’ repair template on top of the three example sequences). Gray rectangles depict ‘random’ sequences not corresponding to the repair-template sequence. The color gradient clarifies to which part of the repair template the insertional fragment corresponds and defines the orientation of the fragment. This is additionally illustrated by arrows overlaying the rectangles. A ‘c’ in the rectangle means that it corresponds with the repair template's complementary sequence. In ‘NT 120 S’ example sequence 3, the integrated repair template fragments could not be assigned to a certain location in the reference sequence, due to the limited length of NGS sequence reads (250 bp), which is depicted by two question marks ('?'). This scheme is a simplified representation. A more detailed scheme of these examples, using the NGS read sequence as reference point, is depicted in Fig. S2.