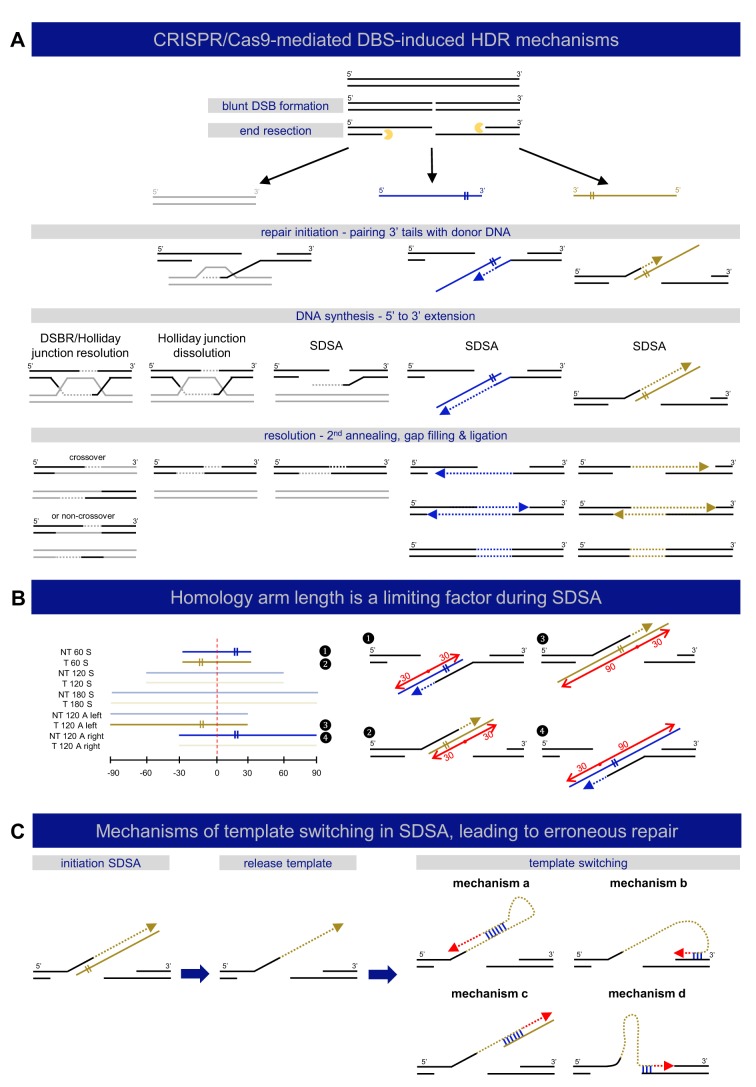
Fig. 6.
Mechanisms of DNA double-strand break repair. (A) A schematic representation of different described mechanisms of DSB-induced HDR is shown, categorized by repair template type: double-stranded (gray) or single-stranded (‘target’ – yellow, ‘non-target’ – blue). DSB repair by HDR is always initiated by DNA end resection, resulting in 3′ single-stranded DNA tails, which participate in strand invasion into homologous sequences. DSB repair with double-stranded repair templates can be carried out by three different mechanisms: double-strand break repair (DSBR or Holliday junction resolution), Holliday junction dissolution, or synthesis-dependent strand annealing (SDSA) (Allers and Lichten, 2001; Szostak et al., 1983). DSB repair using single-stranded templates, sometimes referred to as SSTR, acts via SDSA (Davis and Maizels, 2016; Kan et al., 2017; Paix et al., 2017). (B) Homology-arm length influences the efficiency of SDSA. The left panel depicts the composition of all included repair templates in this study, where the four templates that perform significantly worse in comparison to the others in terms of total repair rates (see Fig. 1B) are highlighted (1-4). The location of the DSB is marked with a red dashed line. These four templates all contain a short (30 nt) homology arm at the side of the repair template that does not anneal to one of 3′ overhangs (marked with ‘II’ sign), but participates in the resolution step, as also depicted in the right panel, where each of these inefficient templates is fitted to the SDSA model for DSB repair with single-stranded templates. The red dots refer to the DSB location, with left and right homology arms marked with red arrows and length indications. (C) Mechanisms of template switching in SDSA leading to erroneous repair. Following SDSA initiation, dissociation of unstable intermediates can lead to template switching. Four possible mechanisms for template switching during SDSA are shown. These mechanisms are based on microhomology (binding marked in blue) and can result in erroneous replication (red dashed arrows).