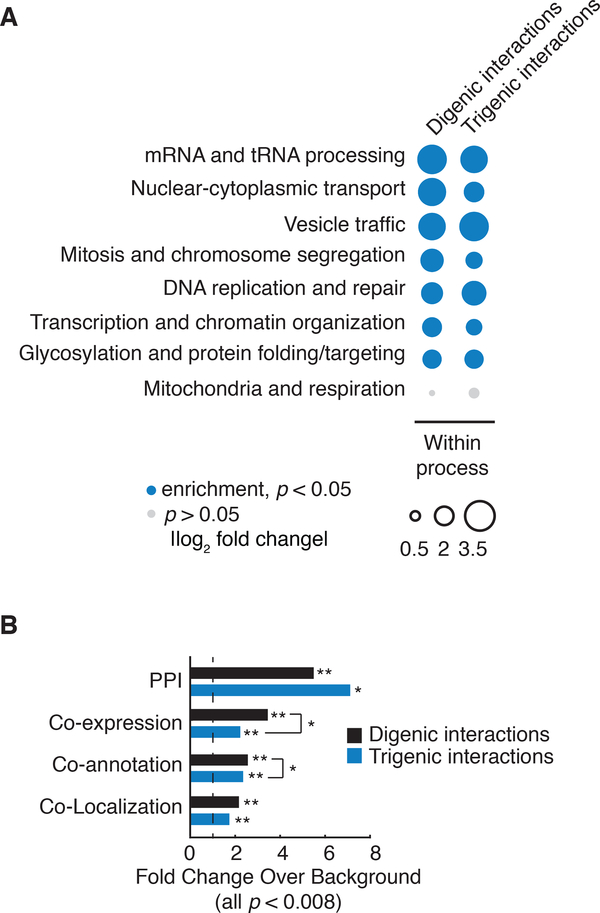
Fig. 2. Functional characterization of trigenic interactions.
(A) Frequency of negative genetic interactions within biological processes. The fraction of screened query-array combinations exhibiting negative interactions belonging to functional gene sets annotated by SAFE on the global genetic interaction network (55). Within process received a count for any combination in which both genes for digenic interactions or all three genes for trigenic interactions, were annotated to the same term. The size of the circle assigned to each process-process element reflects the fold-increase over the background fraction of interactions (digenic = 0.023, trigenic = 0.016). Significance was assessed using hypergeometric cumulative distribution test, p < 0.05. Significant enrichment is shown in filled blue circle and no significant change is in grey. (B) Enrichment of negative digenic (black) and trigenic (blue) interactions across four functional standards. Black dashed line marks no enrichment. The functional standards are the following: merged protein-protein interaction (PPI) standard (56–60), co-annotation is based on SAFE terms (7), co-expression (61), co-localization (62). Significance was assessed using hypergeometric cumulative distribution test, * represents 10−4 ≤ p < 0.01, ** represents p < 10-4.