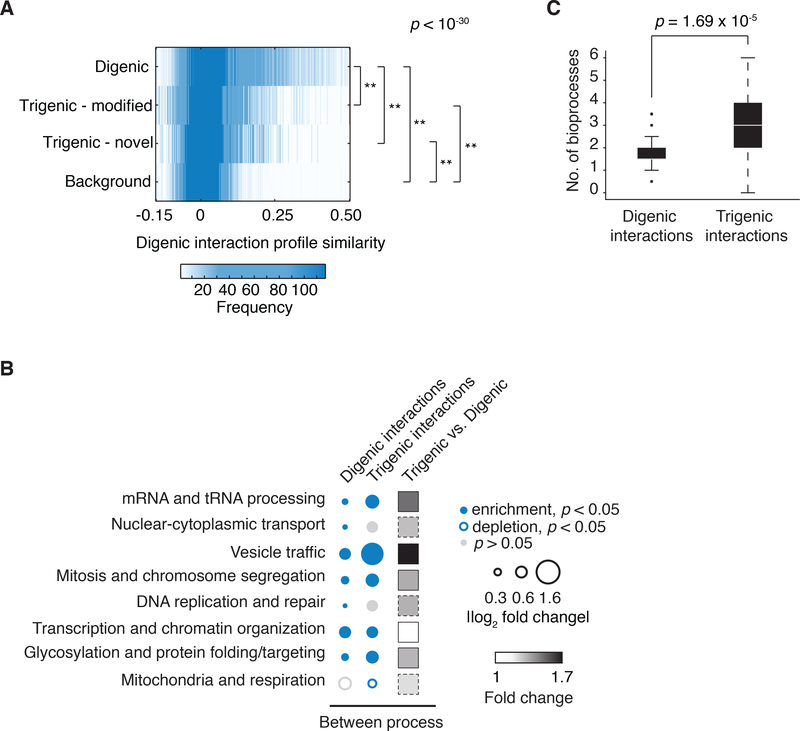
Fig. 6. Trigenic interactions are more functionally distant than digenic interactions.
(A) Distribution of genetic interaction profile similarities of genes showing digenic and trigenic interactions. P-values are based on rank sum median test, all p < 10-34. (B) Frequency of negative genetic interactions between biological processes using SAFE annotations for digenic and trigenic interactions (55). The size of the circle assigned to each process-process element reflects the fold-increase over the background fraction of interactions (digenic = 0.023, trigenic = 0.016), p < 0.05 based on hypergeometric cumulative distribution test. The “between process” category received a count for any combinations that were not counted in the “within process” category shown in Fig. 2A. Significant enrichment is shown in filled blue circle, significant underenrichment in open blue circle, and no significant change is in grey. Trigenic vs. digenic fold change is the ratio of trigenic interaction enrichment to digenic interaction enrichment, and is shown as a filled square (black is maximal fold change, white is no fold change). In cases where the between-process enrichment was observed but is not significant at a p < 0.05, the square is outlined with a dashed line. (C) The number of SAFE bioprocess clusters enriched for digenic or trigenic interactions.