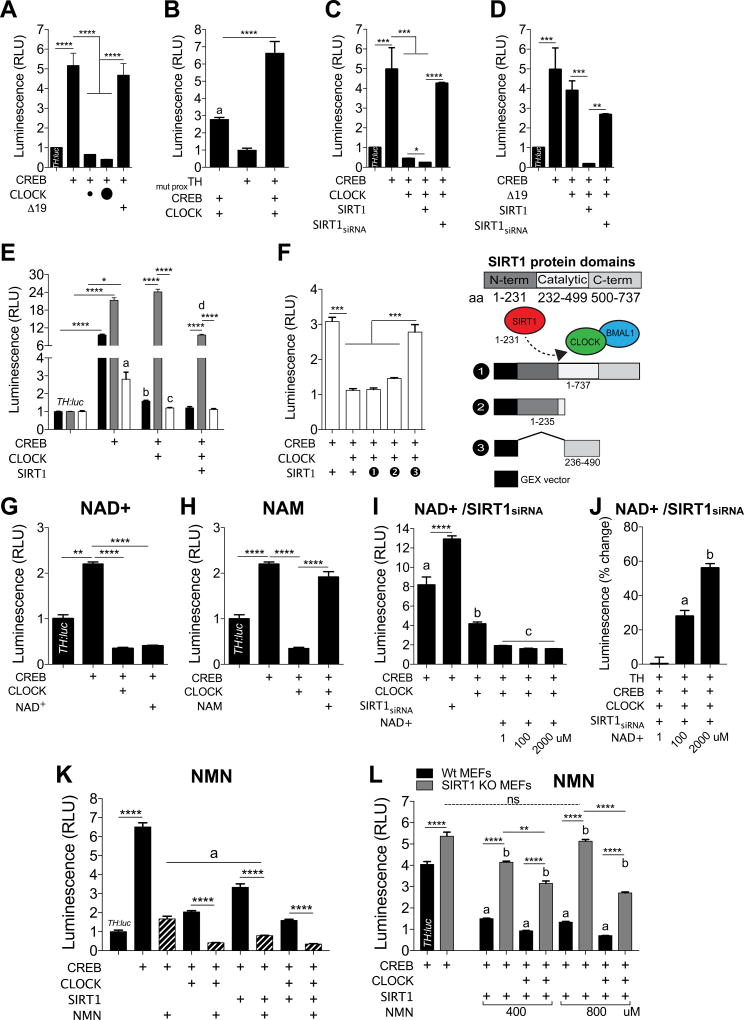
Figure 2. CLOCK, SIRT1, and Metabolic Cofactors Modulate the Transcriptional Activity of TH.
(A) PC12 cells were transfected and TH:luc promoter activity was measured. CREB induces activity of TH:luc, which is prevented by co-transfection with CLOCK, while CLOCKΔ19 fails to do so (one-way ANOVA, F4,30=35.1, p<0.0001; Tukey’s post hoc tests, **** p<0.0001). Amount of CLOCK plasmid transfected (small circle, 1µg; large circle, 10µg). n=8–15 per condition, data represents an average across triplicate wells and multiple plates.
(B) Mutation of the proximal E-box site of the TH promoter prevents the ability of CLOCK to repress transcriptional activity (one-way ANOVA, F7,24=250.3, p<0.0001; Tukey’s post hoc tests, a TH vs. TH/CREB/CLOCK p<0.05, **** p<0.0001). n=4 per condition, data represents an average across triplicate wells and multiple plates.
(C) siRNA-mediated knockdown of SIRT1 prevents the ability of CLOCK to repress transcriptional activity, while SIRT1 further represses activity when co-transfected with CLOCK (one-way ANOVA, F3,32=3503, p<0.0001; Tukey’s post hoc tests, * p<0.05, *** p<0.001, **** p<0.0001). n=4 per condition, data represents an average across triplicate wells and multiple plates.
(D) SIRT1 restores the ability of CLOCKΔ19 to repress transcriptional activity (one-way ANOVA, F2,9=76.5, p<0.0001; Tukey’s post hoc tests, ** p<0.01, *** p<0.001). n=4 per condition, data represents an average across triplicate wells and multiple plates.
(E) TH:luc activity was measured in Wt, SIRT1 KO, and CLOCK KO MEFs. SIRT1 is required for CLOCK to prevent CREB-induced transcriptional activity (genotype × condition, F6,40=3503, p<0.0001). CLOCK represses CREB-induced transcriptional activity in Wt (a TH vs. TH/CREB, p<0.01; b TH/CREB vs. TH/CREB/CLOCK, p<0.0001) and CLOCK KO (c TH/CREB vs. TH/CREB/CLOCK, p<0.0001) MEFs, but fails to do so in SIRT1 KO MEFs. Co-transfection of CLOCK and SIRT1 in CLOCK KO MEFs prevents CREB-induced transcriptional activity (d TH/CREB/CLOCK vs. TH/CREB/CLOCK/SIRT1 p<0.001). n=4–5 per condition, data represents an average across triplicate wells and multiple plates.
(F) CLOCK KO MEFs were transfected with either partial domains or full SIRT1: 1) GEX vector backbone control; 2) N-terminus (required for CLOCK interaction24; or 3) catalytic domain. Full SIRT1 and partial N-terminus SIRT1 prevents CREB-induced TH:luc activity, with no effect of partial catalytic SIRT1 (one-way ANOVA, F4,20=68.14, p<0.0001; Tukey’s post hoc tests, *** p<0.001). n=4–6 per condition, data represents an average across triplicate wells and multiple plates. Transfected construct (+). All data are represented as mean ± SEM.
(G) PC12 cells were treated with NAD+ (1µM), harvested 72 hrs later then measured TH:luc activity. CLOCK and NAD+ prevent CREB-induced TH:luc activity (one-way ANOVA, F8,27=16.23, p<0.0001; Tukey’s post hoc tests, ** p<0.01, **** p<0.0001). n=4 per condition, data represents an average across triplicate wells and multiple plates.
(H) PC12 cells were treated with NAM (10mM), harvested 72 hrs later then measured TH:luc activity. NAM prevents CLOCK-mediated repression of TH:luc activity (one-way ANOVA, F8,27=16.23, p<0.0001; Tukey’s post hoc tests, **** p<0.0001). n=4 per condition, data represents an average across triplicate wells and multiple plates.
(I) PC12 cells were treated with varying concentrations of NAD+, harvested 48 hrs later then measured TH:luc activity. NAD+ further reduces TH:luc activity regardless of dose relative to CLOCK (one-way ANOVA, F6,21=118.1, p<0.0001; Tukey’s post hoc tests, **** p<0.0001, a TH vs. TH/CREB, p<0.0001, b TH/CREB vs. TH/CREB/CLOCK, p<0.05, c TH/CREB/CLOCK vs. TH/CREB/CLOCK + NAD+, p<0.05). n=4 per condition, data represents an average across triplicate wells and multiple plates.
(J) siRNA-mediated knockdown of SIRT1 leads to increased TH:luc activity in the presence of NAD+ (one-way ANOVA, F5,18=67.56, p<0.0001; Tukey’s post hoc tests, a TH/CREB/CLOCK/SIRT1siRNA vs. TH/CREB/CLOCK/SIRT1siRNA + NAD+ (100µM), p<0.05, b TH/CREB/CLOCK/SIRT1siRNA vs. TH/CREB/CLOCK/SIRT1siRNA + NAD+ (2000µM), p<0.05. n=4 per condition, data represents an average across triplicate wells among multiple plates. n=4 per condition, data represents an average across triplicate wells and multiple plates.
(K) PC12 cells were transfected, treated with NMN (200µM), then harvested after 72hrs to measure TH:luc activity. One-way ANOVA followed by Tukey’s multiple comparisons tests, **** p<0.0001. a TH/CREB + NMN vs. TH/CREB/SIRT1 + NMN, p<0.0001. n=6 per condition, data represents an average across triplicate wells among multiple plates. n=6 per condition, data represents an average across triplicate wells and multiple plates.
(L) Transfected Wt and SIRT1 KO MEFs were treated with NMN (400 or 800µM), then harvested 48–72hrs later to TH:luc activity. The repressive effects of NMN on TH:luc activity depended on SIRT1 and CLOCK (genotype × condition, F4,30=50.4, p<0.0001, Tukey’s post hoc tests, ** p<0.01, **** p<0.0001, a condition vs. TH/CREB Wt MEFs, p<0.0001, b condition vs. TH/CREB SIRT1 KO MEFs, p<0.05. n=4 per condition, data represents an average across triplicate wells and multiple plates.
Transfected construct (+). All data are represented as mean ± SEM.