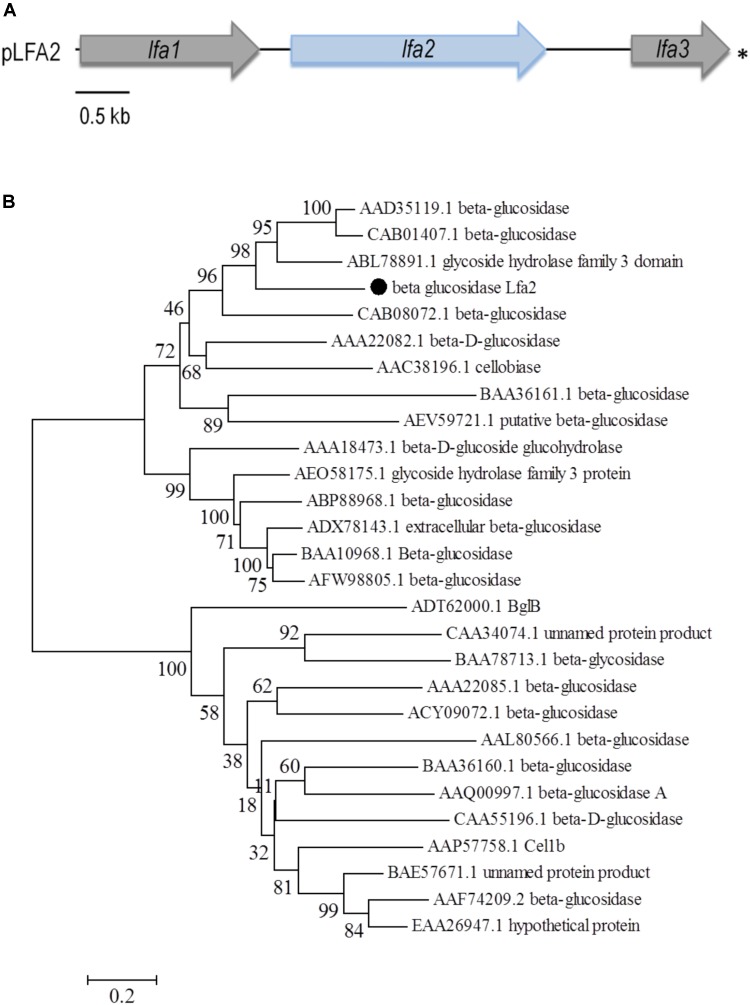
FIGURE 1.
Schematic representation of the ORFs identified in the pLFA2 plasmid and phylogenetic analysis of β-glucosidase Lfa2. (A) The complete metagenomic insert is shown. Arrows display the location, length and transcriptional orientation of the ORFs. Lfa2 ORF is showed as a light blue arrow. Asterisk indicates incomplete ORFs. (B) Phylogenetic tree of 28 candidate sequences using the Neighbor-Joining (NJ) method. Amino acid sequences were recovered from GenBank as indicate by accession numbers as follows: AAD35119.1 (Thermotoga maritima MSB8), CAB01407.1 (Thermotoga neapolitana), ABL78891.1 (Thermofilum pendens Hrk 5), MH397474 (beta glucosidase Lfa2), CAB08072.1 (Clostridium stercorarium), AAA22082.1 (Agrobacterium tumefaciens), AAC38196.1 (Cellulomonas biazotea), BAA36161.1 (Bacillus sp.), AEV59721.1 (uncultured bacterium), AAA18473.1 (Trichoderma reesei), AEO58175.1 (Thermothelomyces thermophila ATCC 42464), ABP88968.1 (Penicillium brasilianum), ADX78143.1 (Aspergillus fumigatus), BAA10968.1 (Aspergillus aculeatus), AFW98805.1 (Aspergillus niger), ADT62000.1 (bacterium enrichment culture), CAA34074.1 (Sulfolobus solfataricus), BAA78713.1 (Thermococcus kodakarensis KOD1), AAA22085.1 (Agrobacterium sp.), ACY09072.1 (uncultured bacterium [2]), AAL80566.1 (Pyrococcus furiosus DSM 3638), BAA36160.1 (Bacillus sp.), AAQ00997.1 (Clostridium cellulovorans), CAA55196.1 (Avena sativa), AAP57758.1 (Trichoderma reesei), BAE57671.1 (Aspergillus oryzae RIB40), AAF74209.2 (Aspergillus niger), EAA26947.1 (Neurospora crassa). Numbers on nodes correspond to percentage bootstrap values for 1,000 replicates.