Fig. 1.
Boxplots of the detection rate, or the proportion of genes in a cell reporting expression values greater 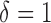 calculated for each cell across 15 publicly available scRNA-seq studies (the number of genes (G) included in each study). Boxplots are ordered by median detection rate across cells within a study. The detection rate across cells and studies ranges from less than 1
calculated for each cell across 15 publicly available scRNA-seq studies (the number of genes (G) included in each study). Boxplots are ordered by median detection rate across cells within a study. The detection rate across cells and studies ranges from less than 1 to 65
to 65 . For Patel and others (2014)
. For Patel and others (2014) , the data submitted to GEO were de-trended so that measurements for each gene averaged to zero across cells and authors applied heavy gene filtering resulting in only
, the data submitted to GEO were de-trended so that measurements for each gene averaged to zero across cells and authors applied heavy gene filtering resulting in only 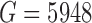 genes. Therefore, the detection rate for this study was calculated by downloading raw sequencing files and quantifying gene-level expression for
genes. Therefore, the detection rate for this study was calculated by downloading raw sequencing files and quantifying gene-level expression for 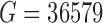 genes. (see Section 2.1 for complete details).
genes. (see Section 2.1 for complete details).

