Table 1.
Column 1 shows the reference. Column 2 shows the organism. Column 3 shows the single-cell protocol used. Column 4 shows the number of cells (samples) included in the study. Column 5 shows the number of genes included in the data uploaded to the public repository with varying gene annotations used. Column 6 indicates the units in which the values were reported. Column 7 shows the level of confounding between biological condition and batch effect quantified using the standardized Pearson contingency coefficient as a measure of association. The percentage ranges from 0
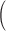 no confounding
no confounding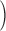 to 100
to 100
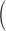 completely confounded
completely confounded
| Study | Organism | scRNA-seq protocol | Number of cells | Number of genes | Processed data available | Confounding ( ) ) |
|---|---|---|---|---|---|---|
| Deng and others (2014) | Mouse | SMART-Seq | 286 | 22 958 | RPKM | 96.6‡ |
| Guo and others (2015) | Human | Tang and others (2009) | 154 | 23 394 | FPKM | 82.1 |
| Kowalczyk and others (2015) | Mouse | SMART-Seq | 533 | 8422 | TPM | 84.8 |
| Kumar and others (2014) | Mouse | SMART-Seq | 361 | 22 443 | TPM | 97.1 |
| Patel and others (2014) | Human | SMART-Seq | 430 | 5948 | TPM | 98.9 |
| Treutlein and others (2014) | Mouse | SMART-Seq | 198 | 23 745 | FPKM | 92.8 |
| Shalek and others (2014) | Mouse | SMART-Seq | 383 | 27 723 | TPM | 100 |
| Trapnell and others (2014) | Human | SMART-Seq | 306 | 47 192 | FPKM | 100 |
| Burns and others (2015) | Mouse | SMART-Seq | 249 | 26 585 | TPM | NA |
| Leng and others (2015) | Human | SMART-Seq | 458 | 19 804 | TPM | NA |
| Jaitin and others (2014) | Mouse | MARS-Seq | 4466 | 20 190 | UMI | NA |
| Macosko and others (2015) | Mouse | Drop-Seq | 49 300 | 16 961 | UMI | NA |
| Satija and others (2015) | Zebrafish | SMART-Seq | 1152 | 13 902 | UMI | NA |
| Zeisel and others (2015) | Mouse | STRT-Seq | 3004 | 19 972 | UMI | NA |
| Zheng and others (2017) | Human | Chromium Single Cell | 20 000 | 27 998 | UMI | NA |
| (10X Genomics) |
The main purpose of this study was to investigate monoallelic gene expression in mouse embryos, but here we consider the different developmental stages (oocyte to blastocyst) as the biological condition as an example
