Abstract
Background: Renal cell carcinoma (RCC) is a common malignancy with high morbidity. MicroRNAs (miRNAs) have been demonstrated to be critical post-transcriptional regulators in tumorigenesis. This study aimed to investigate the effect of miR-543 on the proliferation and metastasis of RCC.
Material and Methods: The expression of miR-543 was examined in clinical samples and RCC cell lines. A498 and 786-O cell lines were employed and transfected with miR-543 inhibitor or miR-543 mimics. The correlation between miR-543 and DKK-1 was determined by luciferase reporter assay. Cell viability and cell cycle were determined by CCK8 and flow cytometry assay. Cell migration and invasion capacity were examined by transwell assay. The protein level of DKK1, β-catenin and pGSK-3β were analyzed by western blotting.
Results: miR-543 was found to be up-regulated in RCC cell lines. Further studies identified DKK-1 as a direct target of miR-543. Moreover, miR-543 overexpression suppressed the expression of DKK-1, and promoted cell proliferation, migration and invasion capacity, while knockdown of miR-543 abrogated above results. MiR-543 knockdown also decreased β-catenin and pGSK-3β levels. In vivo assay verified that miR-543 acts as an oncogene through the regulation of DKK-1 and Wnt/β-catenin signaling pathway.
Conclusion: Our study indicated that miR-543 negatively regulate the expression of DKK-1 in vitro. MiR-543 promotes malignancy phenotypes of RCC both in vitro and in vivo. This regulatory effect of miR-543 may be associated with Wnt/β-catenin signaling pathway. Therefore, miR-543 could be used as a biomarker for predicting the progression of RCC.
Keywords: miR-543, DKK-1, renal cell carcinoma, proliferation, metastasis, Wnt signaling
Introduction
Renal cell carcinoma (RCC) is a common malignancy and accounts for approximately 85% of total renal malignancies 1. The morbidity of RCC has increased rapidly in recent years, and distant metastasis accounts for the majority of cancer-related death in patients with RCC 2. Due to its high resistance to chemotherapy and radiotherapy, the main treatment for localized RCC patients remains radical surgical resection 3, 4. Evidence has suggested that the overall five-year survival rate for patients with distant metastasis were only 10%, showing an unsatisfactory prognosis 5. Therefore, a better understanding of the molecular interactions in the initiation and progression of RCC will be helpful in identifying therapeutic targets or providing new prognostic treatments.
MicroRNAs (miRNAs) are a class of non-coding single-stranded RNA molecules with a length of 20-24 nucleotides 6. These molecules are endogenously synthesized, and they negatively regulate the expression of genes by binding to their 3'-untranslated regions (3'-UTRs) 7. Accumulating literatures have indicated that miRNAs are closely related to various biological processes, and aberrant expression of miRNAs may be associated with tumorigenesis and progression 8. For example, miR-26a regulates the proliferation and metastasis of advanced hepatocellular carcinoma through modulating the c-myc/EZH2 gene 9, miR-543 promotes metastasis of prostate cancer by binding to RKIP 10, and miR-223-3p is down-regulated in osteosarcoma tissues and acts as tumor suppressor by targeting CDH6 11.
Dickkopf proteins belong to one of the four members of extracellular Wnt inhibitors family, which can block signaling by binding to plasma membrane Wnt-receptor complexes 12, 13. It has been reported that DKK-1 can directly bind to Krm1/2 and LRP5/6 transmembrane receptors and subsequently suppress downstream Wnt signaling 14, 15. In our present study, we confirmed that miR-543 functions as an oncogene in RCC cells by promoting cell proliferation, migration and invasion. Furthermore, we reported that miR-543 targets Dickkopf1 (DKK1) and the underlying mechanism of miR-543/DKK1 axis in RCC for the first time, which may be applied to the development of new therapeutic strategies for RCC.
Materials and methods
Clinical specimens
The present study was approved by the Institutional Ethics Committee of Ren min Hospital of Wuhan University and carried out according to the guideline of the ethical management. A total of 38 cases of RCC tissue samples and paired normal samples were collected from the Department of Urology, Ren min Hospital of Wuhan University during 2016-2017. Prior written consent was well informed and signed by all participants. Tumor stage and grade were classified according to the tumor, node, metastasis (TNM) staging system of the American Joint Committee on Cancer (AJCC). All tissues were divided into two parts, with one half fixed in 4% paraformaldehyde, and the other half was immediately frozen and then transferred to -80 °C for later analysis. The clinical data was shown in Table 1.
Table 1.
Relative miR-543 expression and the clinical characteristics of 38 patients with RCC
| MiR-543 expression | |||||
|---|---|---|---|---|---|
| Variable | Group | High | Low | Total | P value |
| Gender | Male | 12 | 9 | 21 | 0.327 |
| Female | 7 | 10 | 17 | ||
| Age | <65 | 13 | 10 | 23 | 0.846 |
| ≥65 | 8 | 7 | 15 | ||
| Histological grade | Low or undiffer | 17 | 7 | 24 | 0.034 |
| Middle or high | 5 | 9 | 14 | ||
| Tumor size | <5 | 8 | 12 | 20 | 0.361 |
| >5 | 10 | 8 | 18 | ||
| Clinical stage | T1-T2 | 9 | 19 | 28 | 0.121 |
| T3-T4 | 6 | 4 | 10 | ||
| Lymph nodes metastasis | Absence | 6 | 10 | 16 | 0.014 |
| Presence | 16 | 6 | 22 | ||
| Distant metastasis | Absence | 8 | 21 | 29 | 0.007 |
| Presence | 7 | 2 | 9 | ||
Cell culture and transfection
Normal human renal proximal tubule epithelial cell line HK-2 and RCC cell lines A498, 786-O, and Caki-2 were purchased from ATCC (American Type Culture Collection, Manassas, VA ,USA) and the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in RPMI 1640 medium (PAN Biotech, Aidenbach, Germany) supplemented with 10% foetal bovine serum (FBS) at 37 °C in a humidified atmospheric conditions of 5% CO2. As for cell transfection, miR-543 mimics and miR-543 inhibitors were constructed by Ribobio Company (Guangzhou, China). Transfections were performed using Lipofectamine 2000 (Invitrogen, CA, USA) according to the manufacturer's protocol. Cells were harvested after 48 hours for further analyses.
Quantitative real-time PCR
Total RNA was extracted from clinical samples and cells using Trizol Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. The RNA purity was was examined using spectrophotometry and 100 ng RNA was reserved for complementary DNA(cDNA) synthesis using reverse transcription kit (ABI, CA) or TaqManH MicroRNA reverse transcription kit (ABI, CA) following the manufacturer's protocol. Real-time quantitative PCR was performed using SYBRH select master mix for CFX (Invitrogen, Carlsbad, CA) and the CFX Connet TM real-time PCR system (Bio-Rad, USA). The expression of miR-543 and DKK1 mRNA was normalized to U6 and β-actin respectively. The relative amount of miRNA or mRNA was calculated via the 2-∆∆Ct method and the primer sequences used are shown in Table 2.
Table 2.
RT-PCR primer sequences
| GENE | Primer sequences (5'-3') |
|---|---|
| DKK1 | F: AGTACTGCGCTAGTCCCACC R: TCCTCAATTTCTCCTCGGAA |
| miR-543 | F: CCAGCTACACTGGGCAGCAGCAATTCATGTTT R: CTCAACTGGTGTCGTGGA |
| U6 snRNA | F: CTCGCTTCGGCAGCACATATACT R: ACGCTTCACGAATTTGCGTGTC |
| GAPDH | F: TGAAGGTCGGTGTGAACGGATTTGGTC R: CATGTAGGCCATGAGGTCCACCAC |
Cell proliferation assay
Cell proliferation of A498 and 786-O cells was measured by cell counting kit-8 (CCK-8) cell viability test kit (Beyotime Institute of Bio-technology, Jiangsu, China). Cells were inoculated into 96-well plates at a density of 2 x 103 cells/well for 24 h, and then cells were stained with 20 μL of CCK-8 reagent for 4 hours after transfection. Cell viability was determined through the absorbance at 450 nm using a spectrophotometer (Thermo Fisher, USA).
Cell cycle assay
Cell cycle was examined by flow cytometry analysis. A498 and 786-O cells were harvested and fixed in 70% ethanol overnight. Then, cells were treated with RNase (50 μg/ml) and PI (50 μg/ml) for 30 min in dark room. Subsequently, cells were subjected to flow cytometer (BD biosciences, USA). Data was analyzed using Cell Quest 3.0 software.
Cell Invasion and Migration Assays
Cell invasion and migration assays were performed using Transwell plates (Corning) with 8-μm-pore size membranes with Matrigel (for invasion assay) or without Matrigel (for migration assay). A498 and 786-O cells were collected 48 h after transfection. Briefly, 3 × 104 cells were seeded in the upper Matrigel-coated chamber, and medium containing 10% FBS was placed in the lower chamber. After incubation at 37˚C for 24 h, cells on the upper chamber membrane were wiped away. Then, cells on the lower chamber membrane were stained with 0.2 % crystal violet for 30 minutes. Five predetermined fields were counted under a microscope (×100). All assays were performed in triplicate.
Plasmid Construction and Luciferase Reporter Assays
The putative and mutated miR-543 target binding sequence in DKK-1 were synthesized and cloned into luciferase reporter to generate the wild-type (DKK-1-WT) or mutated-type (DKK1-MUT) reporter plasmids. The mutant 3'UTR sequence of DKK-1 was obtained using an overlap-extension PCR method. Sequences containing the predicted wild and mutant binding sites were subcloned into a psiCHECK-2 vector (Promega, Madison, USA). For luciferase reporter assay, A498 and 786-O cells were seeded into 24-well plate at a density of 1x105 cells/well. Then, cells were co-transfected with miR-543 mimics or NC vector respectively using Lipofectamine 2000 (Invitrogen, CA, USA) following the instructions of manufacturer. 48 h after transfection, cells were collected and lysed. Luciferase activities were analyzed using a Dual-Luciferase Reporter Assay System (Promega, Madison, USA).
Western blot analysis
Total cellular proteins were lysed using RIPA Buffer (P0013B, Beyotime Institute of Biotechnology), followed by centrifugation at 16,000 rpm for 10 min at 4°C. A bicinchoninic acid assay (BCA; Beyotime, Shanghai, China) was performed to quantify protein concentrations. Briefly, equivalent amounts of protein were resolved by 10% SDS-PACE gels electrophoresis and subsequently blotted onto PVDF membranes followed by blocking and incubation with primary antibodies (anti-DKK1, anti-β-catenin, p-GSK-3β, anti-GSK-3β; Abcam, Cambridge, UK) at 4 °C overnight. GAPDH was used as a loading control. After washing with TBST buffer, membranes were incubated with goat anti-rabbit IgG anti-body at room temperature for 1 hour. All bands were visualized with an ECL system kit (MultiSciences, Hangzhou, China). Optical densities were qualified by ImageJ software (NIH, Bethesda, MD, USA).
Immunohistochemistry
The expression of DKK-1 was examined using immunohistochemical staining. Tissues were fixed in 4% paraformaldehyde, embedded in paraffin and then sliced into 3 μm sections. After that, sections were incubated with primary antibody against DKK-1 (ab61034, Abcam, UK) at a dilution of 1:200 at 4°C overnight. After incubation with secondary antibody, sections were stained with DAB reagent. All sections were photographed at a magnification of × 400.
Tumor xenograft formation assay
Six-week old male BALB/C nude mice were obtained from the Animal Center of Ren'min Hospital of Wuhan University. All animal studies were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and approved by the Animal Care and Use Committee of Wuhan University. A498 Cells transfected with miR-543 mimics or negative control were subcutaneously inoculated into the left flank of nude mice (n=4 per group, 1 × 106 cells for each). Tumor growth was examined every 5 days, and tumor volume was calculated using the equation: Volume = (length × width2) / 2(mm3). 4 weeks after inoculation, animals were sacrificed. Xenografts were dissected and analyzed in subsequent studies.
Statistical Analysis
All statistical analyses were performed using SPSS 19.0 (SPSS Inc, Chicago, IL, USA) and Graphpad Prism 5.0. Data were presented as means ± SD. Differences were assessed by two-tailed Student's t test and χ2 test as appropriate. All experiments were performed at least 3 times. P<0.05 was considered statistically significant.
Results
MiR-543 negatively correlates with the expression of DKK-1
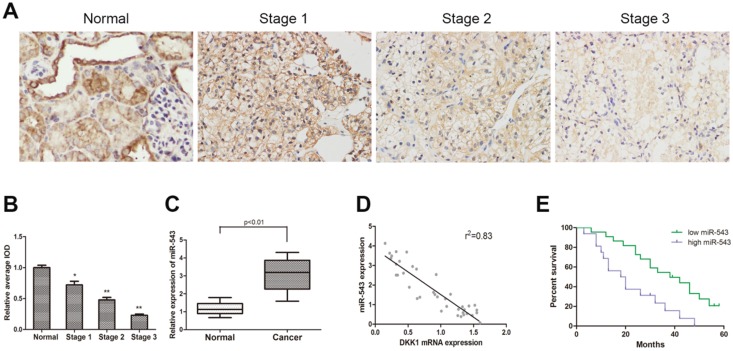
To determine the expression of DKK-1 in RCC tissues and paired adjacent normal renal tissues from 38 RCC patients, immunohistochemistry staining and qRT-PCR were performed. As the result showed, the expression of DKK-1 decreased significantly in RCC tissues compared with normal tissues, and negatively correlated with tumor malignancy grade (Fig 1A). The expression of miR-543 was examined using qRT-PCR. Data suggested that miR-543 was dramatically up-regulated in RCC tissues (Fig 1B). Subsequently, we analyzed clinical data from 38 RCC patients in order to delineate the correlation between miR-543 and clinical features of RCC. All patients were divided into two groups (high miR-543 group and low miR-543 group) according to miR-543 level using ROC curve. Results showed that the expression of miR-543 was closely related to the histological grade, tumor metastasis (Table 1, P<0.05). A Kaplan-Meier analysis and log-rank test suggested that patients with high level miR-543 associated with poor outcome of prognosis (Fig 1D). Subsequently, a two-tailed Pearson's correlation analysis was performed and the result showed that miR-543 was inversely correlated with the expression of DKK-1 (Fig 1C).
Fig 1.
The expression of miR-543 is negatively correlated with DKK1 in clinical RCC specimens. (A) Immunohistochemical staining of DKK1 in RCC tissues and paired normal tissues. (B) Relative expression of DKK1 was detected using average integrated optical density (IOD) *P<0.05, **P<0.01 vs Normal group. (C) Box plot represents relative miR-543 expression levels in RCC tissues and paired normal tissues. (D) A two-tailed Pearson's correlation analysis revealed that the expression of miR-543 is negatively correlated with DKK1. (E) Kaplan-Meier analysis of the correlation between miR-543 expression and overall survival of patients with RCC.
MiR-543 modulates the expression of DKK-1
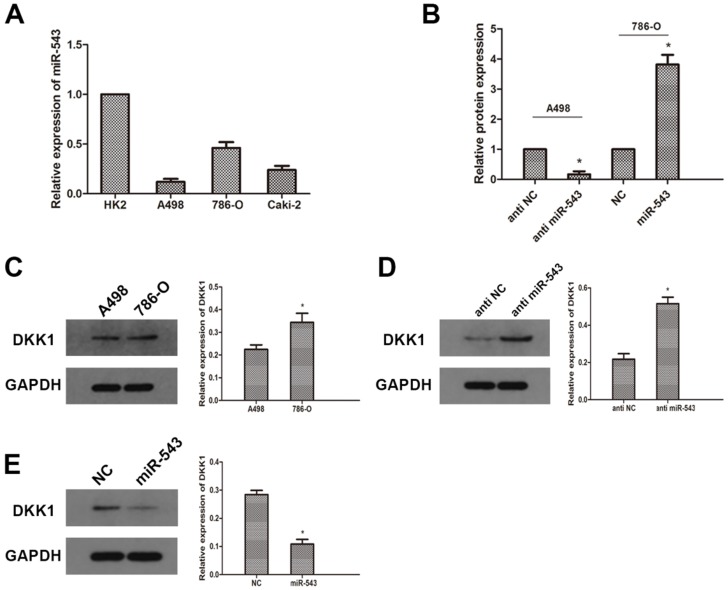
To further investigate the correlation between miR-543 and DKK-1, we examined the expression of miR-543 in four RCC cell lines (A498, 786-O and Caki-2) and human normal renal cell line HK2. As the result of qRT-PCR showed that miR-543 was up-regulated in varying degrees in RCC cells compared with normal cells (Fig 2A). The expression of miR-543 in A498 increased most significantly, while it increased moderately in 786-O cells. Therefore, we employed A498 and 786-O cells in following study. The expression of DKK-1 in these two cell lines was determined using western blot. As the data showed that the expression of DKK-1 in A498 was much less than in 786-O cells (Fig 2C). Then, we transfected A498 cells with miR-543 inhibitors and 786-O cells with miR-543 mimics. Transfection efficiency was examined using qRT-PCR, and the result showed that the expression of miR-543 was significantly suppressed in A498 cells and was overexpressed in 786-O cells (Fig 2B). The expression of DKK-1 was examined after transfection. Results from western blot suggested that DKK-1 was markedly up-regulated in A498 cells transfected with miR-543 inhibitor, while it was moderately weakened in 786-O cells transfected with miR-543 mimics compared with NC groups (Fig 2D, E).
Fig 2.
MiR-543 regulates the expression of DKK1 in RCC cells. The expression of miR-543 in RCC cell lines (A498, 786-O and Caki-2) and human normal renal cell line HK2 was examined by qRT-PCR. *P<0.05 vs. HK2 group. (B) The expression of miR-543 in A498 and 786-O cells transfected with miR-543 mimics and miR-543 inhibitors was measured by qRT-PCR. *P<0.05 vs. respective control group. (C) The expression of DKK1 in A498 and 786-O cells was determined using western blot. *P<0.05 vs. A498 group. (D) Relative expression of DKK1 in A498 cells transfected with miR-543 inhibitors and 786-O cells transfected with miR-543 mimics was examined using western blot. *P<0.05 vs. anti-NC group or NC group.
MiR-543 directly targets DKK-1
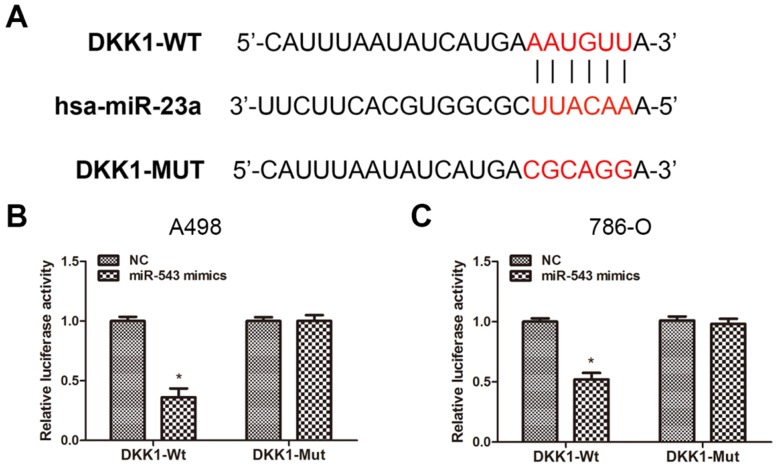
To verify the certain interaction between miR-543 and DKK-1, we employed bioinformatics prediction via open access database (miRwalk 2.0, miRanda and TargetScan). The result showed that DKK-1 contained putative binding sequence for miR-543 in its 3'-UTR (Fig 3A). Therefore, we cloned the putative binding sequence in the 3'UTR of DKK-1 and insert the fragment sequence into a luciferase reporter vector. As the result from luciferase reporter assay revealed, the reporter activity of DKK-1 3'UTR was markedly suppressed upon the overexpression of miR-543 in both two cell lines. On the other hand, the reporter containing mutated DKK-1 3'UTR sequence kept the same activity as NC group (Fig 3B, C). Collectively, above data demonstrated that DKK-1 is a direct downstream target of miR-543.
Fig 3.
DKK1 is a direct target of miR-543. (A) Sequence alignment of predicted miR-543 binding sites within the DKK1 3'UTR and its mutated sequence for luciferase reporter assay. (B, C) Luciferase reporter assay was performed in A498 cells and 786-O cells that were co-transfected with miR-543 mimics and reporter vectors containing DKK1 3'UTR or mutated DKK1 3'UTR. Relative luciferase activities are presented. *P < 0.05 vs. NC group. Data are presented as mean ± SD from three separated experiments.
MiR-543 regulates malignant phenotypes of RCC in vitro
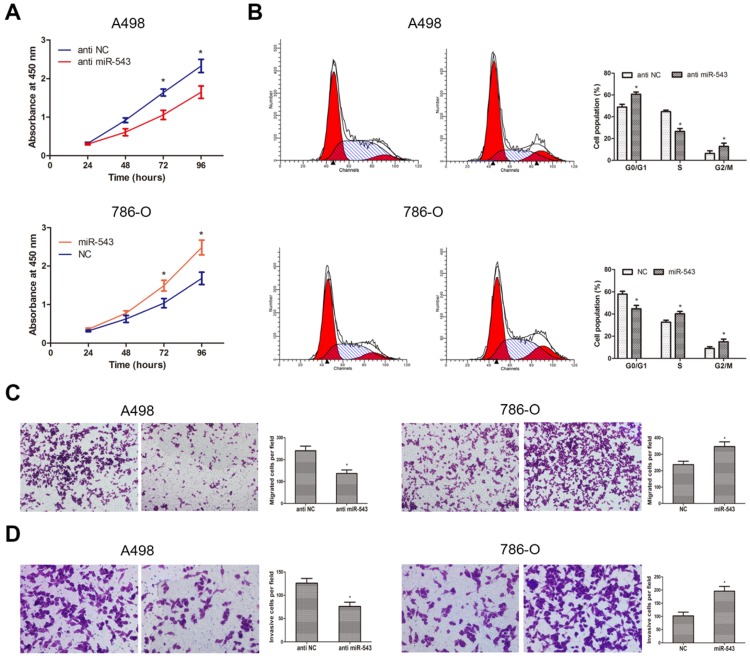
To better understand the biological function of miR-543 in RCC, we explored phenotype changes of RCC cells by gain and loss function assay. Data from CCK-8 assay suggested that miR-543 overexpression markedly promoted cell viability of 786-O cells, while miR-543 knockdown suppressed cell viability of A498 cells (Fig 4A). Subsequently, we determined the effect of miR-543 on cell proliferation using cell cycle assays. As we expected, miR-543 overexpression significantly facilitated cell proliferation of 786-O cells at S phase, while miR-543 knockdown induced cell cycle arrest of A498 cells at G1/G0 phase (Fig 4B). Moreover, we examined the influence of miR-543 on cell migratory and invasive capacities. As results from transwell assays showed, the number of migrated and invaded cells increased obviously in miR-543 overexpression group compared with NC group, while the number markedly decreased in miR-543 knockdown group (Fig 4C, D). Taken together, our study proved that miR-543 functions as an oncogene in RCC cells.
Fig 4.
Influence of miR-543 on the malignant phenotypes of RCC in vitro. (A) CCK-8 assays were performed to determine cellular proliferation in A498 cells transfected with miR-543 inhibitors and 786-O cells transfected with miR-543 mimics. *P<0.05 vs. anti-NC group or NC group. (B) Cell cycle of A498 cells transfected with miR-543 inhibitors and 786-O cells transfected with miR-543 mimics were detected by flow cytometry. *P<0.05 vs. anti-NC group or NC group. (C, D) Transwell invasion assays were performed to examine migratory and invasive ability in A498 and 786-O cells after transfection. Cell number was counted in five random fields at 200 × magnification. *P<0.05 vs. anti-NC group or NC group. Data are presented as mean ± SD from three separated experiments.
MiR-543 modulates RCC progression through Wnt/β-catenin signaling pathway
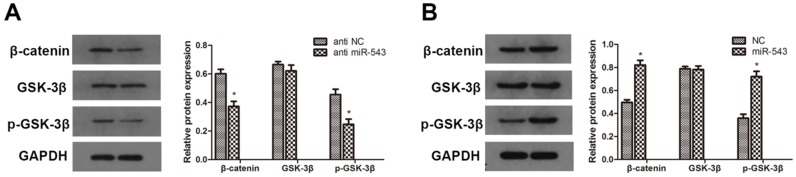
To further investigate whether Wnt/β-catenin signaling pathway involves in the regulation of RCC induced by miR-543, we detected the protein expression of relevant markers using western blot. As the data showed in Fig. 5, the expression of β-catenin and phosphorylated GSK-3β was markedly up-regulated in miR-543 overexpressed 786-O cells (Fig 5A). On the other hand, miR-543 knockdown induced significant inhibition of β-catenin and phosphorylated GSK-3β (Fig 5B). Together, these results demonstrated that Wnt/β-catenin signaling pathway involved in the miR-543 mediated regulatory effect.
Fig 5.
Effects of miR-543 on Wnt/β-catenin signaling pathways. (A, B) The protein levels of β-catenin, p-GSK-3β and GSK-3β in A498 cells transfected with miR-543 inhibitors and 786-O cells transfected with miR-543 mimics were examined by western blot analysis. *P<0.05 vs. anti-NC group or NC group. Data are presented as mean ± SD from three separated experiments.
MiR-543 regulates tumor growth of RCC in vivo
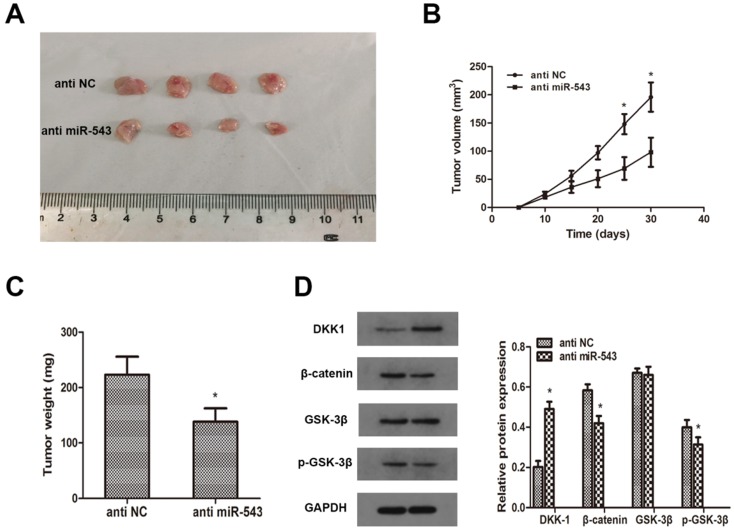
To investigate whether miR-543 induced tumorigenesis and progression of RCC in vivo, A498 cells transfected with miR-543 inhibitors were subcutaneously inoculated into BALB/c nude mice. 4 weeks later after inoculation, xenograft tumors were excised and reserved for further analysis. Tumor volumes and weights were examined, and the results showed that miR-543 knockdown remarkably inhibited tumor growth compared with NC group (Fig 6A, B, C). Subsequently, we determined the expression of DKK-1 in xenografts through western blot. Data revealed that DKK-1 was markedly increased in miR-543 knockdown group than in NC group, while the expression of β-catenin and phosphorylated GSK-3β was significantly suppressed (Fig 6D). Above results suggested that miR-543 may serve as an oncogene in the tumorigenesis and progression of RCC.
Fig 6.
MiR-543 regulates tumor growth of RCC in vivo. (A) Representative images of xenograft tumors excised from nude mice derived 28 days after subcutaneous inoculation of A498 cells transfected with miR-543 inhibitors or anti-NC. (B) Tumor volumes were calculated every 5 days from day 5 to day 28 after inoculation. *P<0.05 vs. anti-NC group, Bars indicate SD. (C) Tumor weight of xenografts were measured and analysed. *P<0.05 vs. anti-NC group, Bars indicate SD. (D) The protein levels of DKK1, β-catenin, p-GSK-3β and GSK-3β in xenografts were examined using western blot. *P<0.05 vs. anti-NC group. Data are presented as mean ± SD in three independent experiments.
Discussion
Despite advances in diagnosis and treatment for RCC patients, the total prognosis of patients with distant metastasis is still not encouraging. Accumulating evidences suggested that nearly one-third of RCC patients have been diagnosed with metastasis at the first visit 16. Moreover, about 40% of RCC patients undergo radical resection surgery relapse or develop a metastasis after a short term remission 17. Hence, it is necessary to uncover the underlying molecular interactions in the progression of RCC and develop more effective therapeutic approaches. A growing body of literatures indicated that miRNAs tightly correlate to tumorigenesis and progression by binding to the mRNA of specific target genes on post-translation level 18. In this situation, we aimed to explore miRNAs that involve in the progression of RCC.
Previous studies have demonstrated that miRNAs modulate the expression of specific genes post-transcriptionally and functions as tumor suppressor or promoter in different tumors 19, 20. Aberrantly expressed miR-543 has been reported to involve in tumor growth and metastasis in various malignancies. Bi et al found that miR-543 promotes proliferation and migration of non-small cell lung cancer by binding to PTEN 21. It has been also revealed that miR-543 acts as an oncogene in osteosarcoma by suppressing PRMT9 22. Our team has reported that miR-543 induced epithelial-mesenchymal transition and proliferation in prostate cancer 10. In our present study, we verified that endogenous expression of miR-543 was remarkably up-regulated in RCC clinical samples and cell lines compared with adjacent normal tissues and normal renal cells. The expression of miR-543 was negatively correlated with DKK-1, suggesting that miR-543 may regulate the tumorigenesis of RCC through DKK-1. Furthermore, our study revealed that miR-543 overexpression significantly promoted cell proliferative, migratory and invasive capacities, while miR-543 knockdown abrogated above influences. Our findings are in accordance with previous reports, suggesting that miR-543 functions like an oncogene in RCC cells and has a potential to be used as a biomarker for diagnosis and prognosis of RCC.
The Wnt/β-catenin signaling pathway is well known for its fundamental mechanisms in cell polarity, cell proliferation and cell fate determination 23, 24. Thus, dysregulation of Wnt signaling is closely linked to tumorigenesis and progression. The triggering of Wnt signaling could prevent GSK-3β from the activation and stabilization of β-catenin, which are strongly associated with tumor metastasis 25, 26. It has been confirmed that there are five Wnt antagonist families including sFRP, Wif1, Xenopus Cerberus, Wise and Dickkopf families 27. Among the four members of Dickkopf family, Dickkopf (DKK)-1 is the first identified Wnt antagonist, which can block wnt/β-catenin signaling by binding to plasma membrane Wnt-receptor complexes 28. Hiroshi et al verified that DKK-1 induced apoptosis and inhibited proliferation of human RCC both in vitro in vivo 29, supporting a tumor suppressor role for this protein. DKK-1 is also found to be downregulated in breast cancer and ovarian cancer 30, 31. In the present study, we predicted that miR-543 directly binds to DKK-1 using bioinformatics methods and then verified such correlation by luciferase reporter assay. Subsequently, we investigated the influence of miR-543 on the expression of DKK-1 and Wnt/β-catenin signaling. Our findings indicated that miR-543 overexpression significantly suppressed the expression of DKK-1 and triggered the activation of β-catenin and phosphorylated GSK-3β. On the other hand, miR-543 knockdown markedly induced DKK-1 up-regulation, suggesting miR-543 acts as an oncogene in the tumorigenesis and progression of RCC, and its carcinogenetic effect may be ascribed to the negative regulation of DKK-1 through Wnt/β-catenin signaling pathway.
Conclusion
In sum, our present study indicated that miR-543 acts as an oncogene in RCC cells by promoting cell proliferation, migration and invasion capacity, and such biological function may, at least in part, be ascribed to the negative regulation of DKK1 through Wnt/β-catenin signaling pathway. These results implied that miR-543 could be used as a diagnostic biomarker for the diagnosis and prognosis of RCC.
Acknowledgments
This study was supported by Wuhan Morning Light Plan of Youth Science and Technology (2017050304010281), Hubei Province Health and Family Planning Scientific Research Project (No. WJ2017M025 and No. WJ2017Z005 ), Natural Science Foundation of Hubei Province (No. 2016CFB114, 2017CFB181) and Research Project of Wuhan University (No. 2042017kf0097).
References
- 1.Jonasch E, Gao J, Rathmell WK. Renal cell carcinoma. BMJ. 2014;349:g4797. doi: 10.1136/bmj.g4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ljungberg B, Campbell SC, Choi HY, Jacqmin D, Lee JE, Weikert S, Kiemeney LA. The epidemiology of renal cell carcinoma. Eur Urol. 2011;60:615–621. doi: 10.1016/j.eururo.2011.06.049. [DOI] [PubMed] [Google Scholar]
- 3.Dutcher JP. Recent developments in the treatment of renal cell carcinoma. Ther Adv Urol. 2013;5:338–353. doi: 10.1177/1756287213505672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Escudier B, Albiges L, Sonpavde G. Optimal management of metastatic renal cell carcinoma: current status. Drugs. 2013;73:427–438. doi: 10.1007/s40265-013-0043-1. [DOI] [PubMed] [Google Scholar]
- 5.Patil S, Ishill N, Deluca J, Motzer RJ. Stage migration and increasing proportion of favorable-prognosis metastatic renal cell carcinoma patients: implications for clinical trial design and interpretation. Cancer-Am Cancer Soc. 2010;116:347–354. doi: 10.1002/cncr.24713. [DOI] [PubMed] [Google Scholar]
- 6.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 7.Kala R, Peek GW, Hardy TM, Tollefsbol TO. MicroRNAs: an emerging science in cancer epigenetics. J Clin Bioinforma. 2013;3:6. doi: 10.1186/2043-9113-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kartha RV, Subramanian S. Competing endogenous RNAs (ceRNAs): new entrants to the intricacies of gene regulation. Front Genet. 2014;5:8. doi: 10.3389/fgene.2014.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X, Zhang X, Wang T, Wang L, Tan Z, Wei W, Yan B, Zhao J, Wu K, Yang A, Zhang R, Jia L. MicroRNA-26a is a key regulon that inhibits progression and metastasis of c-Myc/EZH2 double high advanced hepatocellular carcinoma. Cancer Lett; 2018. [DOI] [PubMed] [Google Scholar]
- 10.Du Y, Liu XH, Zhu HC, Wang L, Ning JZ, Xiao CC. MiR-543 Promotes Proliferation and Epithelial-Mesenchymal Transition in Prostate Cancer via Targeting RKIP. Cell Physiol Biochem. 2017;41:1135–1146. doi: 10.1159/000464120. [DOI] [PubMed] [Google Scholar]
- 11.Ji Q, Xu X, Song Q, Xu Y, Tai Y, Goodman SB, Bi W, Xu M, Jiao S, Maloney WJ, Wang Y. miR-223-3p Inhibits Human Osteosarcoma Metastasis and Progression by Directly Targeting CDH6. Mol Ther; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glinka A, Wu W, Delius H, Monaghan AP, Blumenstock C, Niehrs C. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature. 1998;391:357–362. doi: 10.1038/34848. [DOI] [PubMed] [Google Scholar]
- 13.Niehrs C. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene. 2006;25:7469–7481. doi: 10.1038/sj.onc.1210054. [DOI] [PubMed] [Google Scholar]
- 14.Bafico A, Liu G, Yaniv A, Gazit A, Aaronson SA. Novel mechanism of Wnt signalling inhibition mediated by Dickkopf-1 interaction with LRP6/Arrow. Nat Cell Biol. 2001;3:683–686. doi: 10.1038/35083081. [DOI] [PubMed] [Google Scholar]
- 15.Causeret F, Sumia I, Pierani A. Kremen1 and Dickkopf1 control cell survival in a Wnt-independent manner. Cell Death Differ. 2016;23:323–332. doi: 10.1038/cdd.2015.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chatzizacharias NA, Rosich-Medina A, Dajani K, Harper S, Huguet E, Liau SS, Praseedom RK, Jah A. Surgical management of hepato-pancreatic metastasis from renal cell carcinoma. World J Gastrointest Oncol. 2017;9:70–77. doi: 10.4251/wjgo.v9.i2.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ko JJ, Xie W, Kroeger N, Lee JL, Rini BI, Knox JJ, Bjarnason GA, Srinivas S, Pal SK, Yuasa T, Smoragiewicz M, Donskov F, Kanesvaran R, Wood L, Ernst DS, Agarwal N, Vaishampayan UN, Rha SY, Choueiri TK, Heng DY. The International Metastatic Renal Cell Carcinoma Database Consortium model as a prognostic tool in patients with metastatic renal cell carcinoma previously treated with first-line targeted therapy: a population-based study. Lancet Oncol. 2015;16:293–300. doi: 10.1016/S1470-2045(14)71222-7. [DOI] [PubMed] [Google Scholar]
- 18.Skaftnesmo KO, Prestegarden L, Micklem DR, Lorens JB. MicroRNAs in tumorigenesis. Curr Pharm Biotechnol. 2007;8:320–325. doi: 10.2174/138920107783018390. [DOI] [PubMed] [Google Scholar]
- 19.Okato A, Goto Y, Kurozumi A, Kato M, Kojima S, Matsushita R, Yonemori M, Miyamoto K, Ichikawa T, Seki N. Direct regulation of LAMP1 by tumor-suppressive microRNA-320a in prostate cancer. Int J Oncol. 2016;49:111–122. doi: 10.3892/ijo.2016.3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin Y, Liu AY, Fan C, Zheng H, Li Y, Zhang C, Wu S, Yu D, Huang Z, Liu F, Luo Q, Yang CJ, Ouyang G. MicroRNA-33b Inhibits Breast Cancer Metastasis by Targeting HMGA2, SALL4 and Twist1. Sci Rep. 2015;5:9995. doi: 10.1038/srep09995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bi M, Chen W, Yu H, Wang J, Ding F, Tang DJ, Tang C. miR-543 is up-regulated in gefitinib-resistant non-small cell lung cancer and promotes cell proliferation and invasion via phosphatase and tensin homolog. Biochem Biophys Res Commun. 2016;480:369–374. doi: 10.1016/j.bbrc.2016.10.055. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H, Guo X, Feng X, Wang T, Hu Z, Que X, Tian Q, Zhu T, Guo G, Huang W, Li X. MiRNA-543 promotes osteosarcoma cell proliferation and glycolysis by partially suppressing PRMT9 and stabilizing HIF-1alpha protein. Oncotarget. 2017;8:2342–2355. doi: 10.18632/oncotarget.13672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 24.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 25.Fukumoto S, Hsieh CM, Maemura K, Layne MD, Yet SF, Lee KH, Matsui T, Rosenzweig A, Taylor WG, Rubin JS, Perrella MA, Lee ME. Akt participation in the Wnt signaling pathway through Dishevelled. J Biol Chem. 2001;276:17479–17483. doi: 10.1074/jbc.C000880200. [DOI] [PubMed] [Google Scholar]
- 26.Zhou BP, Deng J, Xia W, Xu J, Li YM, Gunduz M, Hung MC. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol. 2004;6:931–940. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
- 27.Kawano Y, Kypta R. Secreted antagonists of the Wnt signalling pathway. J Cell Sci. 2003;116:2627–2634. doi: 10.1242/jcs.00623. [DOI] [PubMed] [Google Scholar]
- 28.Niehrs C. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene. 2006;25:7469–7481. doi: 10.1038/sj.onc.1210054. [DOI] [PubMed] [Google Scholar]
- 29.Hirata H, Hinoda Y, Nakajima K, Kawamoto K, Kikuno N, Ueno K, Yamamura S, Zaman MS, Khatri G, Chen Y, Saini S, Majid S, Deng G, Ishii N, Dahiya R. Wnt antagonist DKK1 acts as a tumor suppressor gene that induces apoptosis and inhibits proliferation in human renal cell carcinoma. Int J Cancer. 2011;128:1793–1803. doi: 10.1002/ijc.25507. [DOI] [PubMed] [Google Scholar]
- 30.Lv C, Li F, Li X, Tian Y, Zhang Y, Sheng X, Song Y, Meng Q, Yuan S, Luan L, Andl T, Feng X, Jiao B, Xu M, Plikus MV, Dai X, Lengner C, Cui W, Ren F, Shuai J, Millar SE, Yu Z. MiR-31 promotes mammary stem cell expansion and breast tumorigenesis by suppressing Wnt signaling antagonists. Nat Commun. 2017;8:1036. doi: 10.1038/s41467-017-01059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen MW, Yang ST, Chien MH, Hua KT, Wu CJ, Hsiao SM, Lin H, Hsiao M, Su JL, Wei LH. The STAT3-miRNA-92-Wnt Signaling Pathway Regulates Spheroid Formation and Malignant Progression in Ovarian Cancer. Cancer Res. 2017;77:1955–1967. doi: 10.1158/0008-5472.CAN-16-1115. [DOI] [PubMed] [Google Scholar]