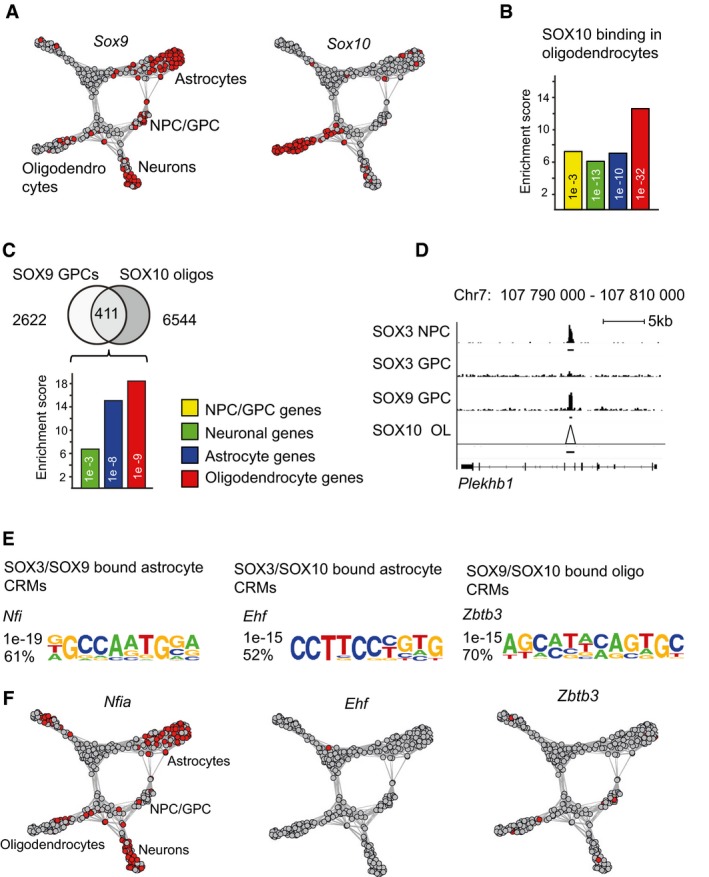
Graphical view showing SOX9 and SOX10 expression among spinal cord cells analyzed with scRNA‐seq.
Bar graph shows enrichment of genes bound by SOX10 in rat oligodendrocytes within the differentially expressed gene sets. Yellow bar represents enrichment within NPC/GPC genes, green bar within neuronal genes, blue bar within astrocytic genes, and red bar within oligodendrocytic genes. SOX10 ChIP‐seq data from Ref.
25.
Venn diagram shows 2,622 SOX9‐bound sites in GPCs, which are not co‐targeted by SOX3, and their overlap with SOX10‐bound sites in mature oligodendrocytes. Bar graph shows that mostly oligodendrocyte‐specific genes are sequentially bound by SOX9 in GPCs and by SOX10 in oligodendrocytes. However, SOX9 and SOX10 also bind genes that are selectively expressed in astrocytes.
ChIP‐seq peak graphics around the oligodendrocyte gene Plekhb1. ChIP‐seq peaks are derived from four different experiments: SOX3 ChIP‐seq in NPCs, SOX3 ChIP‐seq in GPCs, SOX9 ChIP‐seq in GPCs, and SOX10 ChIP‐seq in oligodendrocytes. Both ChIP‐seq reads and called peak regions (underlying black lines) are shown for all data sets except for SOX10, for which only called peak regions are shown (black line). Triangle in the SOX10 data set indicates called peak. SOX10 ChIP raw reads could not be transferred from rn5 to mm9 assembly, and therefore, only SOX10 peak regions are shown in the figure. Scale bar: 5 kb.
Figure shows most strongly enriched DNA‐binding motifs within three specific peak sets: SOX3‐ and SOX9‐bound regions around astrocyte‐specific genes, SOX9‐ and SOX10‐bound regions around astrocyte‐specific genes, and SOX9‐ and SOX10‐bound regions around oligodendrocyte‐specific genes.
Expression of Nfia, Ehf, and Zbtb3 among spinal cord cells analyzed with scRNA‐seq. Red color indicates cells with RPKM values above 55 for Nfia, 5 for Zbtb3, and 5 for Ehf. n = 3 for SOX9 and 2 for SOX3. P‐Values (phyper, R) are calculated from the total number of genes for mm10 (23,389).