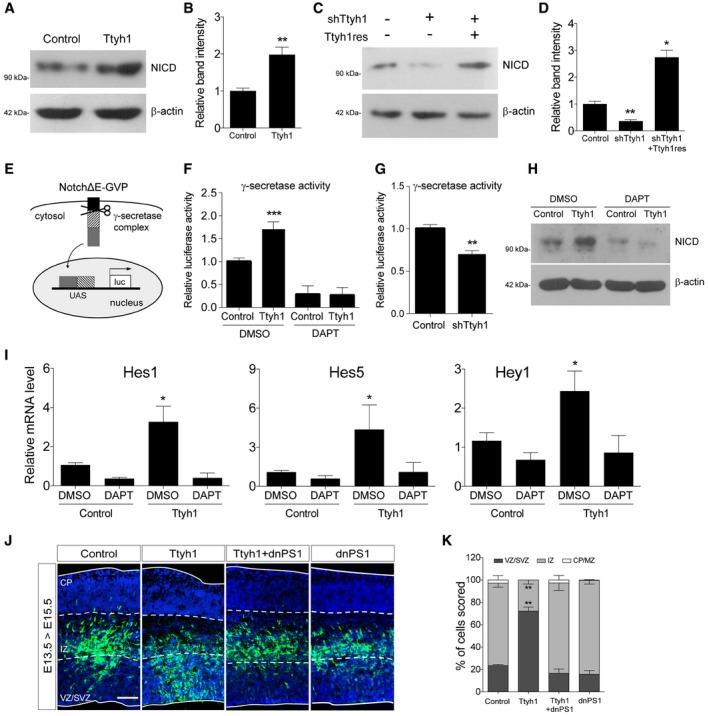
-
A–D
Effects of Ttyh1 expression (A) or Ttyh1 knockdown (C) on the generation of NICD at 2 days posttransduction of E14.5 neural progenitor cells were analyzed by Western blotting using an anti‐NICD antibody. (B, D) Quantification of band intensities in (A, C) (n = 4 for B, n = 3 for D).
-
E
Schematic representation of the γ‐secretase reporter assay. Following cleavage at the γ‐secretase site in NotchΔE‐Gal4‐VP16 (NotchΔE‐GVP), the intracellular domain containing the GVP domain translocates into the nucleus and triggers luciferase expression driven by the UAS promoter (Figure redrawn from Karlström
et al
31).
-
F, G
(F) Ttyh1‐transduced E14.5 neural progenitors were incubated with DAPT for 2 days, then cells were harvested and lysates were used for luciferase assay (n = 3). (G) Effects of Ttyh1 knockdown on γ‐secretase activity were also assessed using E14.5 neural progenitor cells (n = 4 for F, n = 3 for G).
-
H, I
Ttyh1‐transduced E14.5 neural progenitors were incubated for 2 days in the presence or absence of DAPT, and samples were prepared for Western blotting using anti‐NICD antibody (H) or qPCR for Notch target genes (I) (n = 3).
-
J, K
(J) The indicated plasmids were electroporated into E13.5 brains, and brain samples were harvested at E15.5 for anti‐GFP immunolabeling. GFP and DAPI signals are shown in green and blue, respectively. (K) Quantification of GFP+ cell localization in (J) (n = 3). dnPS1, dominant negative presenilin 1; VZ, ventricular zone; SVZ, subventricular zone; IZ, intermediate zone; CP, cortical plate. Scale bar: 100 μm.
Data information: Error bars represent SD. Student's
‐test was used to determine statistical significance. *
0.001.