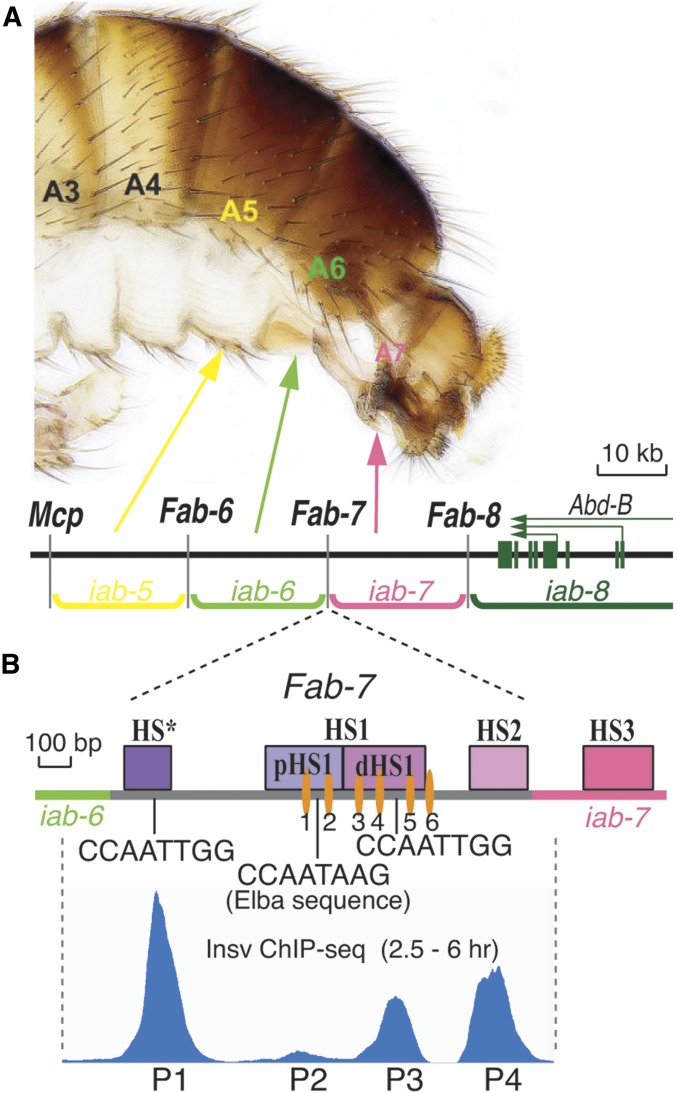
Figure 1.
The Fab-7 boundary. (A) Map of the Abd-B region of the bithorax complex. The relative location of the Abd-B regulatory domains—iab-5, iab-6, iab-7, and iab-8—and the segments that they specify in the adult male fly are indicated in the panel. Also shown are the positions of the boundary elements—Mcp, Fab-6, Fab-7, and Fab-8—and the Abd-B transcription unit. (B) Map of the Fab-7 nuclease hypersensitive sites—HS*, HS1, and HS2—and location of the binding motifs (GAGAG) for the GAGA factor (GAF) (orange ovals) and Insv. Also shown is a diagram of the Insv chromatin immunoprecipitation (ChIP)-sequencing peaks spanning the Fab-7 boundary and the adjacent iab-7 PRE (polycomb response element), adapted from Dai et al. (2015). The peaks are aligned with position of the Fab-7 hypersensitive sites.