Abstract
Anthocyanin accumulation specifically depends on sucrose (Suc) signaling. However, the molecular basis of this process remains unknown. In this study, in vitro pull-down assays identified ETHYLENE-INSENSITIVE3 (EIN3), a component of both sugar signaling or/and metabolism. This protein interacted with YDA, and the physiological relevance of this interaction was confirmed by in planta co-immunoprecipitation, yeast two-hybrid (Y2H) assay, and bimolecular fluorescence complementation. Ethylene insensitive3-like 1 (eil1) ein3 double-mutant seedlings, but not ein3-1 seedlings, showed anthocyanin accumulation. Furthermore, ein3-1 suppressed anthocyanin accumulation in yda-1 plants. Thus, EMB71/YDA-EIN3-EIL1 may form a sugar-mediated gene cascade integral to the regulation of anthocyanin accumulation. Moreover, the EMB71/YDA-EIN3-EIL1 gene cascade module directly targeted the promoter of Transparent Testa 8 (TT8) by direct EIN3 binding. Collectively, our data inferred a molecular model where the signaling cascade of the YDA-EIN3-TT8 appeared to target TT8 via EIN3, thereby modulating Suc signaling–mediated anthocyanin accumulation.
Keywords: EMB71/YODA(YDA), anthocyanin biosynthesis, ETHYLENE-INSENSITIVE3 (EIN3), TT8, sugar signal or/and metabolism, ethylene signal
ANTHOCYANINS are a class of flavonoids widely found in plants and play important roles in many of physiological processes, including functioning as photoprotective screens in vegetative tissue, visual attractors in pollination, antimicrobial agents, and feeding deterrents in the defense response (Winkel-Shirley 2001).
Anthocyanin accumulation is stimulated via many endogenous signals, for example, sucrose (Teng et al. 2005), auxin, abscisic acid (ABA) (Jeong et al. 2004, 2010; Hoth et al. 2010), gibberellin (Weiss et al. 1995), cytokinin (Deikman and Hammer 1995), jasmonates (Qi et al. 2011), and ethylene (Morgan and Drew 1997). Further, the synthesis of these molecules is also induced by a series of environmental stresses, including attempted microbial infection, ultraviolet irradiation, insect attack, wounding, nutrient depletion, and drought (Qi et al. 2011).
MYB/bHLH/TTG1 (MBW) components are thought to regulate anthocyanin biosynthesis in a spatiotemporal manner (Teng et al. 2005). In Arabidopsis thaliana, the WD-repeat protein Transparent Testa Glabra 1 (TTG1) (Walker et al. 1999) recruits basic helix-loop-helix (bHLH) transcription factors (Toledo-Ortiz et al. 2003); for example, Transparent Testa 8 (TT8) (Nesi et al. 2000; Baudry et al. 2006), Glabra 3 (GL3) (Payne et al. 2000), or Enhancer of Glabra 3 (EGL3) (Zhang et al. 2003). Further, R2R3 MYB transcription factors (MYB113, MYB90, MYB114, or MYB75) (Borevitz et al. 2000; Stracke et al. 2001, 2007; Zimmermann et al. 2004; Gonzalez et al. 2008; Rowan et al. 2009) can also be recruited. Collectively, these proteins form the WD-repeat/bHLH/MYB component complex. This complex regulates anthocyanin biosynthesis via promoting the expression of late anthocyanin biosynthetic genes, including leucoanthocyanidin dioxygenase, NADPH-dependent dihydroflavonol reductase (DFR), and UDP-Glc:flavonoid 3-Oglucosyltransferase (UF3GT) (Dooner et al. 1991; Kubasek et al. 1992; Shirley et al. 1995; Gonzalez et al. 2008).
Sucrose (Suc) is a well-characterized endogenous developmental signal. Sugars (e.g., Suc) play dual functions as transported carbohydrates in vascular plants and as signal molecules that regulate gene expression and plant development. Sugar-mediated signals indicate carbohydrate availability and regulate metabolism by coordinating sugar production and mobilization with sugar usage and storage (Baier et al. 2004). Anthocyanin biosynthesis is modulated by changes in Suc concentrations and the sugar-dependent upregulation of the anthocyanin synthesis pathway is Suc-specific (Solfanelli et al. 2006; Jeong et al. 2010). The Arabidopsis pho3 mutant is defective in Suc transporter 2 (SUC2) function. SUC2 encodes a phloem-loading Suc-proton symporter, enabling accumulation of soluble sugars; thus, pho3 plants exhibit growth retardation and also anthocyanin accumulation (Lloyd and Zakhleniuk 2004). Microarray analysis of mature leaves from pho3 plants revealed an increased expression of PRODUCTION OF ANTHOCYANIN PIGMENT 1 (PAP1), PRODUCTION OF ANTHOCYANIN PIGMENT 2 (PAP2), and TT8 transcription factors, as well as of genes encoding anthocyanin biosynthesis enzymes, implying that sugar is a trigger of anthocyanin biosynthesis in vivo (Lloyd and Zakhleniuk 2004).
YODA (YDA), a mitogen-activated protein kinase kinase kinase (MAPKKK), is a member of the MEKK subfamily of the MEKK1/Ste11/Bck1 class of MAPKKKs and is a key component of the regulatory pathways for both embryo and stomatal development (Bergmann et al. 2004; Lukowitz et al. 2004). Current reports indicate that YDA is involved in the regulation of floral patterning (Bemis et al. 2013), drought tolerance (Meng and Yao 2015), anthocyanin accumulation (Meng et al. 2016), the accumulation of reactive oxygen intermediates, and cell death development (Li et al. 2015). A previously reported microarray analysis of yda mutants (Lukowitz et al. 2004) indicated that only 14 out of 8000 genes had changes in expression levels of at least twofold; this gene set included those associated with sugar metabolism or carbohydrate metabolic process (AT5G57550, AT2G43570, and AT4G15760), and cell wall synthesis (At2g45220). Thus, over half were related to sugar metabolism and/or sugar signaling. A further microarray study of yda plants revealed that over 11% of upregulated genes were involved in cell wall differentiation (Bergmann et al. 2004), consistent with the cell walls of differentiated epidermal cells being extensively reinforced. Collectively, these results suggest that YDA might be involved in sugar metabolism and/or sugar signaling. However, to date, there are no reports providing data to support this hypothesis.
While there are currently many different signaling pathways identified as influencing anthocyanin biosynthesis in Arabidopsis, it is fully unknown how these signal pathways are integrated. Here, we found that loss of YDA function enhanced anthocyanin accumulation and conveyed insensitivity to sugar signaling or/and metabolism. Further, YDA was found to interact with EIN3 in vitro and in vivo put-down, which were confirmed by yeast two-hybrid (YH2) and bimolecular fluorescence complementation (BiFC). Thus, EMB71/YDA-EIN3-EIL1 may form a sugar-regulated gene cascade that controls anthocyanin accumulation. Interestingly, the YDA-EIN3/EIL1 complex directly targets to the TT8 promoter for transcriptional repression, thereby negatively regulating anthocyanin biosynthesis. Our data suggests this is mediated via direct EIN3 binding to the TT8 promoter, repressing the expression of this transcriptional activator. Collectively, our data suggests that the YDA-EIN3-TT8 gene cascade modules may target the TT8 promoter by direct EIN3 binding, regulating anthocyanin biosynthesis.
Materials and Methods
Plant materials and growth conditions
The yda-1 and yda-2 (Lukowitz et al. 2004), ein3-1, eil1-3, ctr1-1 and ein3/eil1 (An et al. 2010) mutants, and the estradiol-inducible EIN3–FLAG transgene (An et al. 2010) within a Col-0 background were all described previously.
The yda-1, yda-2, ctr1-1, tt8-1, and emb71 mutants were obtained from the Arabidopsis Biological Resource Center (Ohio State University). The EIN3-FLAG, eil1-3, ein3-1, and ein3/eil1 seeds were kindly provided by Prof H. W. Guo (Peking University, China). The pHB-YDA:GFP plasmid was kindly provided by Prof. H. Q Yang (Shanghai JiaoTong University, China). The pHB-35Spro-YDA:GFP transgene is driven by the 35S promoter (Bergmann et al. 2004; Kang et al. 2009).
The yda-1/+ein3 mutant was obtained from F2 seedlings of a yda-1/+ein3-1 cross that had shortened roots (Lukowitz et al. 2004) and had lengthened hypocotyls and roots on solid Murashige and Skoog Medium (MS) medium with 6.0 μm l-aminocyclopropane-l-carboxylic acid (ACC) (An et al. 2010). The yda/ein3 mutant was identified from offspring of yda-1/+ein3-1 that had clustered stomata on the cotyledon in the dark (Kang et al. 2009), and then homozygous lines were identified through PCR-based genotyping for ein3-1. Primers for ein3-1 were described previously (An et al. 2010). Similarly, we obtained ein3/tt8 and ein3/eil1/tt8 mutants. The method of double-mutant construction has been described by Kang et al. (2009).
For simultaneous germination, seeds were treated with jarovization at 4° overnight and then sown on solid MS medium supplemented with 1% sucrose (pH 5.8) and 0.8% agar (Meng and Yao 2015). Seedlings grown on agar were maintained in a growth room under 16:8 hr of light/dark cycle with cool white fluorescent light at 21 ± 2° (Meng and Yao 2015). Plants grown in soil were maintained in a controlled environment growth chamber under 16:8 hr light/dark cycle with cool white fluorescent light at 21 ± 2° (Meng and Yao 2015).
Confocal laser scanning microscope for GFP or YFP imaging
Subcellular localization of relative fusion protein/gene expression was assayed within the abaxial epidermis of cotyledons in 1-week-old transgenic plants harboring the relative vector. An Olympus IX-70 microscope (http://www.olympus-global.com/) was used to detect GFP or YFP expression. The sections were photographed under a confocal laser scanning microscope.
Y2H assays
The Y2H analysis was carried out using a GAL4-based Y2H system from MatchmakerGold Systems (Clontech, Palo Alto, CA). The full-length complementary DNA (cDNA) of YDA was cut with NotI and SfiI and ligated into pGBKT7 to construct a bait plasmid. The full-length cDNA of EIN3 was cloned into pGADT7 to construct prey plasmid using BamHI and XhoI. Primers for pGADT7+EIN3 were YTH-EIN3-F: CGC GGA TCC ATG ATG TTT AAT GAG ATG GGA (BamH), and YTH-EIN3-R: CCG CTC GAG TTA GAA CCA TAT GGA TAC ATC (XhoI); and primers for pGBKT7-YDA were YTH-YDA-F: ATG GCC ATG GAG GCC ATG CCT TGG TGG AGT AAA TCA (SfiI), and YTH-YDA-R: ATA AGA ATG CGG CCG CTT AGG GTC CTC TGT TTG TTG (NotI). To test the interaction, the bait plasmid and the prey plasmid were cotransformed into yeast strain Y2H Gold (Clontech) and yeast cells containing both vectors were selected using synthetic dropout (SD)/-Leu/-Trp (DDO) medium. The transformants were assayed by plating the transformed cells onto the SD/-Ade/-His/-Leu/-Trp selective medium (QDO) and QDO/Aba/X-α-Gal selective medium using at least 10 independent colonies for the β-galactosidase test. Serial dilutions of cotransformed yeast cells were used to measure the strength of the interaction.
BiFC assays
To perform this assay, pBI121-YFPC was produced by using YFPC-F (K): 5′ CGG GGT ACC TAC CCA TAC GAT GTT CCA GAT T 3′ KpnI and YFPC-R (S): 5′ CGA GCT CTT ACT TGT ACA GCT CGT CCA TG 3′ ScaI; and pBI121-YFPN was produced by using YFPN-F (K): 5′ CGG GGT ACC GAG CAA AAG TTG ATT TCT GAG G 3′ KpnI and YFPN-R (S): 5′ CGA GCT CTT AGG CCA TGA TAT AGA CGT TGT 3′ ScaI; and pBI121-YFPN-EIN3 was produced by using YTH-EIN3-F: CGC GGA TCC ATG ATG TTT AAT GAG ATG GGA BamHI and YTH-EIN3-R: CCG CTC GAG TTA GAA CCA TAT GGA TAC ATC XhoI; and pBI121-YFPC-YDA was produced by using BIFC-YDA-F: GGG GTA CCA TGG AAA AAA GGG AGA TTG C KpnI and BIFC-YDA-R: CCG CTC GAG AAA GCA TGA TCC AAA AGC TG XhoI. To perform the BiFC assays, leaf blades of Nicotiana benthamiana were transformed transiently with Agrobacterium tumefaciens. The Agrobacterium strains containing the BiFC constructs were grown on OD = 1.0, spun down, and then resuspended in infiltration buffer (10 mM 2-(4-Morpholino) ethanesulfonic acid (MES), 10 mM MgCl2, and 100 mM acetosyringone) to OD = 1.3. Then, they were incubated on a shaker at room temperature for 3–4 hr. Strains containing the two assayed constructs were mixed and then injected into the middle of leaf blades. Leaf blades were imaged via laser scanning confocal microscope and DIC to 3 days after injection. Every interacting pair was tested for two repeated experiments.
Assaying seedling response to high concentration of sugar and ethylene
The use of 1 and 5% mannitol, sucrose and glucose was performed, as has been shown by Meng et al. (2016). Ethylene, ACC, and AgNO3 conditions were described by Jeong et al. (2010).
Assay of sugar metabolites
Twelve-day-old seedlings were ground in liquid nitrogen, and then the powder was isolated with 1 ml of 80% ethanol for 1 hr. These extracts were centrifuged with 12,000 × g for 10–15 min. The supernatant along with ethanol buffer was transferred to a fresh tube and evaporated under vacuum for dryness for 40–60 min. The residues were then redissolved in 600 μl of double-distilled water and kept at 70° for 10–15 min.
Using chloroform:isoamyl alcohol (24:1, v/v), the above aqueous fraction was extracted twice to three times before HPLC analysis. Sugars were quantified and identified via chromatography on an Agilent carbohydrate column and tested with a refractive index detector (Altex 156; Altex Scientific Inc.). Concentrations were measured from peak heights using sucrose, fructose, and glucose (20 mg/ml) as standard samples. These experiments were repeated at least twice, with similar results.
Measurement of anthocyanins
For anthocyanin extraction, the given samples were placed in 50 ml of 1% HCl in methanol (v/v) and then incubated overnight in the dark at 4° with gentle shaking. After this process, 300 ml of water and 300 ml of chloroform were added and mixed to the extract. After centrifugation at 12,000 rpm for 2 min, the absorbance of the supernatant was measured at 530 and 657 nm and the concentration of anthocyanin was determined by using A530- 0.25A657 (Rabino and Mancinelli 1986). These experiments were repeated at least two times with similar results.
Quantitative PCR
Total RNA was extracted from tissues indicated in the figures by the TRIzol reagent (Invitrogen, Carlsbad, CA), as has been described by Meng and Yao (2015) and Meng et al. (2018). SYBR green was used to monitor the kinetics of PCR product in real-time RT-PCR, as has been described by Meng and Yao (2015). Primers of TT8, PAP2, and MYBL2 have been described by Jeong et al. (2010). These experiments were repeated at least two times, with similar results.
Protein expression and purification
The plasmids pGEX-5X-1 (for EIN3, CINV2, SUC1, and HXK1) and pET28a (for YDA) was utilized. The coding sequence of YDA was amplified by the primer pair (5′-GC-GGCCTTTTTGGCC-ATGCCTTGGTGGAGTAAATCAA-3′ and 5′-ATAAGAAT-GCGGCCGC-TTAGGGTCCTCTGTTTGTTGAT-3′) and cloned into the Not1 and Sfi1 restriction sites of pET28a. The coding sequence of CINV2 was amplified by the primer pair (5′-CGGGATCC-TGGAGGAAGGTCATAAAGAAC-3′ and 5-GGAATTCTCAGCAAGTCCATGAAGCAGAT-3′) and cloned into the Bam1H and EcoR1 restriction sites of pGEX-5X-1.
The coding sequence of EIN3 was amplified by the primer pair (5′-GGATCC ATGATGTTTA ATGAGATGGG-3′ and 5′-CTCGAGTGCTCTGTTTGGGAT-3′) and cloned into the Bam1H and XhoI restriction sites of pGEX-5X-1. The coding sequence of HXK1 was amplified by the primer pair (5′-GGATCCATGGGTAAAGTAGCTGTTGGA-3′ and 5′-CTCGAGTTAAGAGTCTTCAAGGTAGAG-3′) and cloned into the Bam1H and XhoI restriction sites of pGEX-5X-1. The coding sequence of SUC1 was amplified by the primer pair (5′-GAATTC ATGGGAGCCTATGAAACAGA-3′ and 5′-CCCGGGCTAGTGGAATCCTCCCATGGT-3′) and cloned into the Bam1H and SmaI restriction sites of pGEX-5X-1. Recombinant glutathione S-transferase binding protein (GST)–tagged EIN3, CINV2, HXK1, and SUC1 and recombinant histidine (HIS) binding protein–tagged YDA were extracted from transformed Escherichia coli (Rosetta2) after 10 hr of incubation at 16°, following induction with 10 μM isopropyl ß-D-1-thiogalactopyranoside. These recombinant proteins were purified using HIS or GST agarose affinity, respectively.
Chromatin immunoprecipitation PCR
The transgenic lines containing estradiol-inducible EIN3-FLAG and 35S:EIN3-GFP were utilized. Chromatin immunoprecipitation (ChIP) was performed with seedlings (Meng 2015). Leaf blades were incubated in buffer [1.0 mM PMSF, 0.5 M sucrose, 1 mM ethylenediaminetetraacetic acid (EDTA), 10–12 mM Tris (pH 8.0), and 1% formaldehyde] under vacuum for 15–20 min for cross-linking the chromatin. Then, 0.1 M Gly was placed in the mixture, incubating for an additional 5 min for terminating the reaction. Leaf blades were placed and ground in liquid nitrogen and resuspended to lysis buffer [150 mM NaCl,1 mMEDTA, 0.1% SDS, 0.1% deoxycholate, 50 mM HEPES (pH 7.5), 1% Triton X-100, 10 mM sodium butyrate, 1 mM PMSF, and 13 complete protease inhibitor; Roche]. Chromatin was sheared to ∼200–500 bp fragments via sonication followed by centrifuged. At 4°, supernatants were precleared under protein G agarose beads for 1–1.5 hr. Input material (supernatant containing chromatin) was used for immunoprecipitation with anti-FLAG antibody and anti-GFP antibody. Anti-FLAG antibody and anti-GFP antibody bound to EIN3-FLAG or GFP-chromatin complexes was incubated with protein G agarose beads for 1–1.5 hr at 4–6°, and then washed several times and eluted with elution buffer. Input and immunoprecipitated chromatin were uncross-linked for 6 hr at 6° with 5 M NaCl. Immunoprecipitated chromatin and input were used for PCR analysis The ChIP DNA products were analyzed PCR using five pairs of primers that were synthesized to amplify ∼300 bp DNA fragments in the promoter region of TT8, PAP2, and MYBL2. Primer sequences (F-5′-atgttcagtttagctttagttcg-3′ and R-5′-aaacttaaatatataacttgaatttg-3′)-TT8-T1, (F-5′-tagaattcaatgcggctggaatat-3′ and R-5′-taaatttaattaattcaattagtatttg-3′)-TT8-T2, and (F-5′-catccaacgtctggtgaacc-3′ and R-5′- tcttagtgcttttgcaacatgtt-3′)-TT8-T3 were used for TT8. Primer sequences (F-5′-cttcggaaaaactgaccggtt-3′ and R-5′-tttcatatccagagaatattccg-3′)-PAP2-P1, (F-5′-gtatttggttcgattcaaacatgc-3′ and R-5′-ggtcttttaatattattcgacaaa-3′)-PAP2-P2, and (F-5′-aaagtttttgtaagaaggtgcac-3′ and R-5′-tatatatatacacgtgaagggc-3′)-PAP2-P3 were used for PAP2. Primer sequences (F-5′-acattcatgattccacaaattctaa-3′ and R-5′-ctttttcaaagtgagattggttg-3′)-MYBL2-M1 and (F-5′-ATGAACAAAACCCGCCTTCG-3′ and R-5′-GGGTCGATTCCCATTTTTACG-3′)-MYBL2-CDS were used for MYBL2. These experiments were repeated at least three times, with similar results.
In vitro pull-down assay
HIS-YDA, GST-EIN3, GST-CINV2, GST-SUC1, and GST-HXK1 expression constructs were prepared as described by Li et al. (2002). The potential for in vitro interaction between YDA and these GST fusion proteins was tested. Briefly, the HIS-YDA fusion and Ni-nitrilotriacetic acid sefinose resin (Sangon, Shanghai, China) were mixed at 4° for 2 hr of rocking, followed by brief centrifugation to precipitate beads and then washed three to four times with PBS buffer supplemented with 0.5% Tween 20. The precipitated beads were mixed with GST fusion protein or GST in the in vitro binding buffer (50 mM Tris-HCl, pH 7.5, 0.15 M NaCl, 1 mM DTT, 0.5% Triton X-100, and 1 mM phenylmethylsulfonyl fluoride) and incubated at 4° for 2 hr of rocking, followed by brief centrifugation to precipitate beads and then washed three to four times with PBS buffer supplemented with 0.5% Tween 20. The bound proteins were resolved by SDS-PAGE for immunoblot analysis using an anti-GST antibody. The reactive bands were visualized via exposure to nitroblue tetrazolium/bromochloroindolyl phosphate. These experiments were repeated at least four times, with similar results.
Co-immunoprecipitation analysis
Co-immunoprecipitation experiments using wild-type and transgenic plant extracts were performed according to Li et al. (2002) with minor modifications. Transgenic plants harboring both FLAG-EIN3 and GFP-YDA expression constructs were harvested. Soluble protein extracts were obtained using a protein extract kit (Sangon). Protein extracts were mixed with anti-FLAG M2 magnetic beads (Sigma, St. Louis, MO) and incubated at 4° overnight with gentle rocking, followed by brief centrifugation to precipitate beads and then washed three to four times with PBS buffer supplemented with 0.5% Tween 20. Co-immunoprecipitated GFP-YDA was screened by western analysis with anti-GFP (Sigma) antibodies. These experiments were repeated at least three times, with similar results.
Electrophoresis mobility shift assay
The electrophoresis mobility shift assay (EMSA) was performed using the LightShift Chemiluminescent EMSA Kit (20148; Pierce) according to the manufacturer’s instructions. The biotin-labeled TT8-T1 DNA fragments (5′-attcaaaaatcaaaagtcaacttttaatgcacttgagttttggtcatgcattcatatacatacatata-3′) and its mutated PTT8-T1 DNA fragments (5′-attctttaatcaaaagtcaacttttaatgcacttgagttttggtcatgcattcatatacatacatata-3′), and the biotin-labeled TT8-T2 DNA fragments (5′-aacattcaaatataattagaggaccgtccaattggtatatgatcctactatattatttgggaagtacatttt-3′) and its mutated PTT8-T2 DNA fragments (5′-aacattcttttataattagaggaccgtccaattggtatatgatcctactatattatttgggaagtacatttt-3′) were synthesized, annealed, and used as probes. The corresponding biotin-unlabeled DNA fragments were utilized as competitor sequences in the assay. The probes were incubated with EIN3 fusion protein at room temperature for 20 min in a binding buffer [56 concentrations: 50 mM HEPES-KOH (pH 7.5), 375 mM KCl, 6.25 mM MgCl, 1 mM DTT, 0.5 mg/mL BSA, glycerol 25%]. Each 20 ml binding reaction contained 25 fmol Biotin-probe, 6 mg protein, and 1 mg poly(deoxyinosinic-deoxycytidylic) acid to minimize nonspecific interactions. The reaction products were analyzed by 6.5% native polyacrylamide gel electrophoresis. This experimental process has been described previously (Meng et al. 2015a,b).
Data availability
The authors affirm that all data necessary for confirming the conclusions of the article are present within the article and figures. Strains and/or plasmids are available upon request. Supplemental figures are available in supporting figures and legends file. Supplemental Material, File S1 shows that HXK1, SUC1, and CINV2 cannot directly interacts with YDA in vitro. File S2 shows that PAP2 and MYBL2 are not target genes of EIN3. All genomic data and genomic sequencing raw reads are available online at https://www.arabidopsis.org/. Supplemental material available at Figshare: https://doi.org/10.6084/m9.figshare.6974570.
Results
yda mutants are not defective in ethylene signaling
We recently reported that YDA can induce anthocyanin accumulation by regulating sucrose levels (Meng et al. 2016). Anthocyanin biosynthesis is negatively regulated through ethylene signaling and positively controlled through sugar signaling (Joeng et al. 2010). An antagonistic interaction between glucose and ethylene signaling has been widely observed (Yanagisawa et al. 2003). Given that YDA appears to regulate anthocyanin accumulation by a sugar signaling pathway, we explored if YDA might also regulate anthocyanin accumulation through ethylene function.
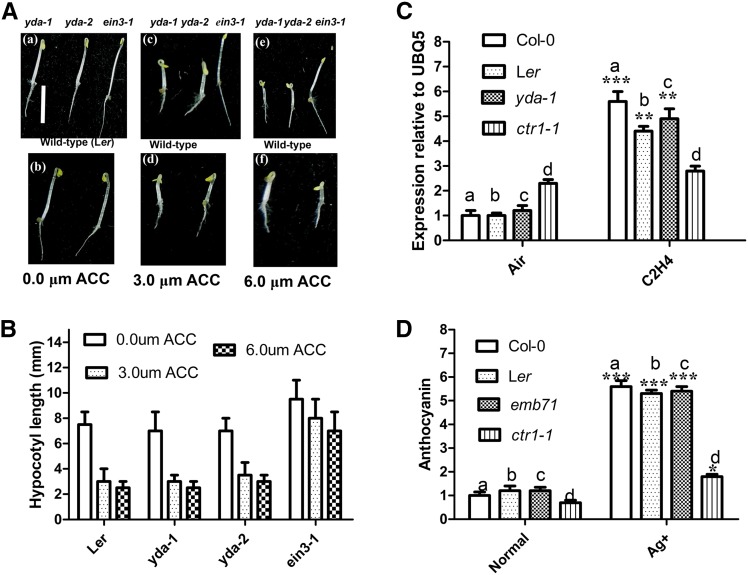
Using the methods of An et al. (2010), statistical analysis of hypocotyl and root growth showed that yda mutants produced both shortened hypocotyls and roots upon ACC treatment, similar to wild-type Ler seedlings [Figure 1, A (a)–(f) and B]. Consistent with these phenotypes, the expression level of ethylene response gene ERF1 was similar between wild-type and yda-1 seedlings. However, the gene was expressed to high levels in the constitutive triple response 1 (ctr1) mutant, which exhibited continuous ethylene signaling (Figure 1C). The inhibition of ethylene signaling by silver enhances the anthocyanin content of corn seedlings and wild-type Arabidopsis seedlings (Jeong et al. 2010). Treatment with silver resulted in significantly enhanced anthocyanin accumulation in both wild-type and yda seedlings but not in ctr1-1 seedlings (Figure 1D). Consistent with these findings, a previous study (Lukowitz et al. 2004) also proposed that yda-1 and yda-2 do not affect ethylene signaling. Taken together, our data suggested that YDA may not be required for ethylene function.
Figure 1.
EMB71/YDA is not involved in ethylene signaling. (A) Representative 4-day-old yda-1 (a), yda-2 (a), ein3-1 (a), and Ler (b) seedlings grown on solid MS medium without ACC (1-aminocyclopropane-1-carboxylic acid); 4-day-old yda-1 (c), yda-2 (c), ein3-1 (c), and Ler (d) seedlings grown on solid MS medium with 3.0 m ACC; and 4-day-old yda-1 (e), yda-2 (e), ein3-1 (e), and Ler (f) seedlings grown on solid MS medium with 6.0 m ACC. Bar, 5.0 mm for a–f. (B) Bar graph exhibiting the difference in hypocotyl length between Ler, yda-1, yda-2, and ein3-1 seedlings grown on solid MS medium with 0.0 m ACC, 3.0 mACC, and 6.0 mACC, respectively. Error bars represent SD (n = 16). Experiments were repeated three times with similar results. (C) Bar graph exhibiting the difference of expression of ERF1 between 8-day-old wild-type (Col-0), Ler, yda-1, and ctr1-1 light-grown seedlings with or without ethylene (25 ppm) for 5 hr. Data were from quantitative RT-PCR. Error bars indicate SD (n = 3; ** P < 0.01, *** P < 0.001). Value of wild-type seedlings without ethylene is set at 1.0. Quantifications were normalized to the expression of UBQ5. (D) Bar graph exhibiting the difference in anthocyanin accumulation between 10-day-old wild-type (Col-0), Ler, emb71, and ctr1-1 seedlings grown under white light. Wild-type value is set as 1.0. Error bars represent SD (n = 14; * P < 0.05, *** P < 0.001). Experiments were repeated two times with similar results.
YDA interacts with EIN3 in vitro and in vivo
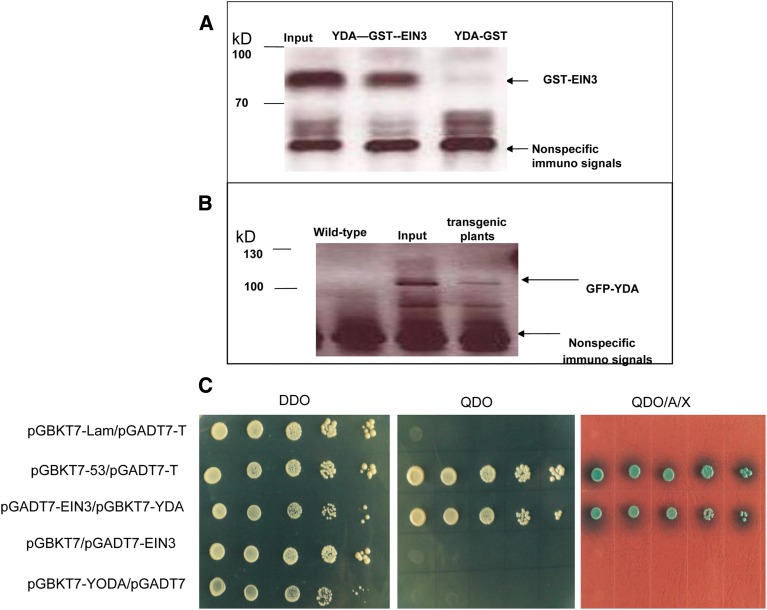
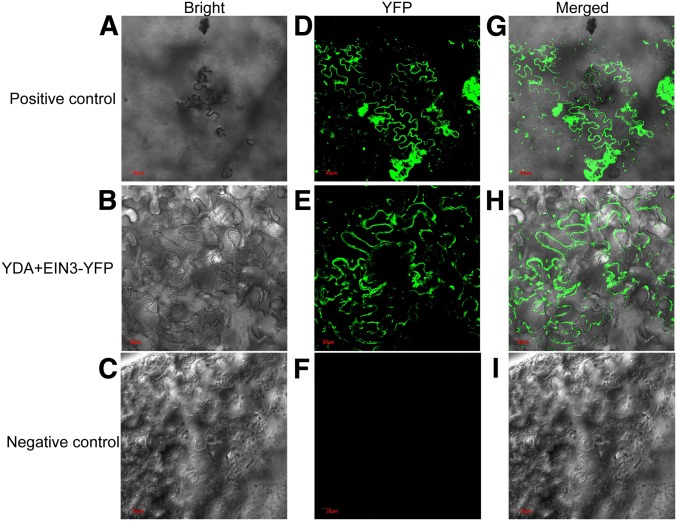
Key genes integral to sugar metabolism and/or signaling include EIN3, HXOKINASE1 (HXK1), SUCROSE-PROTON SYMPORTER1 (SUC1), and CYTOSOLIC INVERTASE2 (CINV2) (Yanagisawa et al. 2003; Barratt et al. 2009). As YDA belongs to the MEKK1/Ste11/Bck1 class of MAPKKKs, which regulate protein targets by phosphorylation, YDA might regulate one or more of EIN3, HXK1, SUC1, and CINV2 at a post-translational level. To identify the potential target(s) of EMB71/YDA in the context of sugar signal transduction, we performed in vitro pull-down experiments. Full-length YDA was expressed as a HIS fusion protein, and full-length EIN3, HXK1, SUC1, and CINV2 were expressed as GST fusion proteins. Following mixing of the fusion proteins, sefinose resin was used to bind selectively to the HIS-YDA fusion protein. The potential presence of the coprecipitated GST fusion protein was examined using a GST antibody. Results indicated that YDA bound specifically to EIN3 in vitro (Figure 2A) but not to HXK1, SUC1, or CINV2 (Figure S1). Moreover, we performed in vivo co-immunoprecipitation experiments. Following transformation and associated genetic crosses, transgenic plants expressing both GFP-YDA and FLAG-EIN3 were produced. Subsequently, the total soluble protein in wild-type and FLAG-EIN3 transgenic seedlings was isolated, and an anti-FLAG antibody was utilized to potentially immunoprecipitate FLAG-EIN3. Potentially co-immunoprecipitated GFP-YDA was detected using a GFP antibody. Our data revealed a direct interaction between YDA and EIN3 in planta (Figure 2B). To confirm that YDA bound specifically to EIN3, we performed YH2 assay and BiFC. Our findings indicated that YDA could specifically interact with EIN3 (Figure 2C and Figure 3). Therefore, a YDA-EIN3 interaction may be relevant for the regulation of anthocyanin accumulation.
Figure 2.
YDA interacts with EIN3. (A) HIS-YDA fusing protein exhibited specific affinity for GST-EIN3 but not GST in vitro. Input is considered as a positive control. (B) GFP-YDA showed specific affinity with FLAG-EIN3 in vivo. FLAG-EIN3 was associated with membranes and can be detected with anti-GFP antibodies. Wild type was used as a negative control. Input is considered as a positive control. (C) The cDNA of YODA and EIN3 were respectively inserted into pGBKT7 and pGADT7 to obtain the bait plasmid and the prey plasmids. Saccharomyces cerevisiae strain Y2H gold was cotransformed with pGBKT7-YODA and pGADT7-EIN3 vectors, and transformed Y2H Gold cells containing both vectors were selected using SD/-Leu-Trp(DDO), SD/-Leu-Trp-His-Ade(QDO), and SD/-Leu-Trp-His-Ade with X-α-gal and Aba(QDO/A/X). pGBKT7 and pGADT7 were bait and prey vectors without inserts. pGBKT7-53 and pGADT7-T encode two fusion proteins that are known to interact (Clontech). pGBKT7-Lam and pGADT7-T were used as negative controls. Serial dilutions of cotransformed yeast cells were used to measure the strength of the interaction.
Figure 3.
BiFC-based interactions between YDA and EIN3. Interactions with (A–C) bright, (D–F) YFP, and (G–I) merged. Pairs of proteins of YDA and EIN3 fused with halves of the YFP molecule were transiently expressed in wild tobacco leaves and reconstituted YFP fluorescence was imaged in epidermis on the abaxial leaf blades. Only EIN3 fused with halves of the YFP molecule was used as a negative control. Plasmid containing YFP was used as a positive control. Bar, 40 μm for A, D, and G; bar, 20.0 μm for B, C, E, F, H, and I.
Increase in anthocyanin accumulation in ein3-1, eil1-3, and ein3/eil1 mutants is related to endogenous sucrose accumulation
Increased anthocyanin accumulation in yda mutants is closely related to enhanced endogenous sucrose accumulation (Meng et al. 2016), and YDA interacts with EIN3 in vitro and in vivo (Figure 2 and Figure 3). Therefore, whether the increase in anthocyanin accumulation in ein3-1, eil1-3, and ein3/eil1 mutants is related to endogenous sucrose accumulation should be determined.
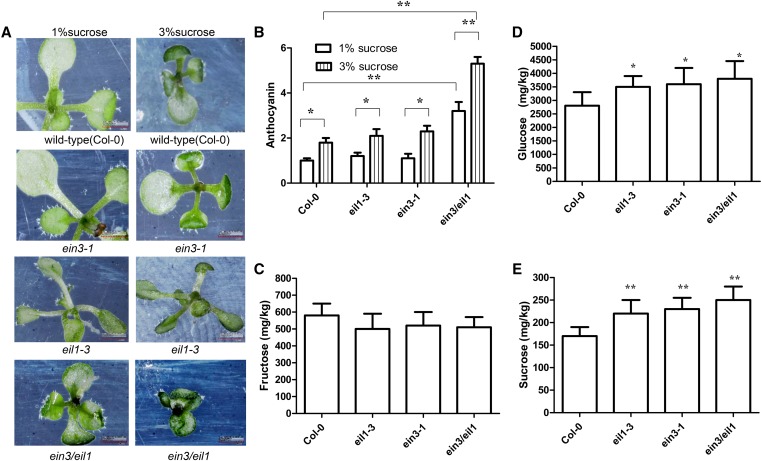
The Arabidopsis genome contains five EIN3 paralogs (EIL1–5). Among these paralogs, EIN3 and EIL1 are the most closely related. EIL1 expression complements the ein3 mutant and constitutively activates the ethylene response pathway (Chao et al. 1997). Thus, EIN3 and EIL1 appear redundant in biological function. When grown on solid MS medium supplemented with both 1% sucrose and 3% sucrose, no significant difference in anthocyanin accumulation was observed on the epidermis of the abaxial leaf surface between wild-type plants and the partial ethylene-insensitive mutants ein3-1 and eil1-3 (Figure 4, A and B). However, ein3/eil1 plants presented increased anthocyanin accumulation compared with wild type, ein3-1, and eil1-3 (Figure 4, A and B). Given that the endogenous sugar concentration can influence anthocyanin accumulation, we determined sucrose, glucose, and fructose levels in ein3-1, eil1-3, and ein3/eil1 plants. Our findings indicated that the concentrations of sucrose and glucose, but not that of fructose, were significantly higher in ein3-1, eil1-3, and ein3/eil1 than in wild-type seedlings (Figure 4, C–E).
Figure 4.
Alternation of anthocyanin accumulation in ein3-1 and ein3/eil1 mutants under abnormal endogenous sucrose accumulation. (A) Ten-day-old wild-type, eil1-3, ein3-1, and ein3/eil1 seedlings grown under long light (16 hr light/8 hr dark) conditions on MS medium supplemented with 1% sucrose and 3% sucrose, respectively. Magnifications are the same. Seedlings were converted for showing abaxial side of leaves and then were photographed. (B) Bar graph exhibiting the difference in the anthocyanin accumulation under A. Col-0 is set as 1.0. Error bars represent SD (n = 3; ** P < 0.01). (C–E) Bar graphs exhibiting the different concentrations of hexose (glucose and fructose) (C and D) and sucrose (E) between wild-type (Col-0), eil1-3, ein3-1, and ein3/eil1 seedlings grown under long light (16 hr light/8 hr dark) conditions on MS medium supplemented with 1% sucrose, respectively. Error bars represent SD (n = 3; * P < 0.05, ** P < 0.01).
When grown on solid MS medium supplemented with either 1 or 3% sucrose, increased anthocyanin accumulation was observed within the epidermis of the abaxial surface on ein3/eil1 leaves relative to the corresponding organs in wild-type, eil1-3, and ein3-1 seedlings (Figure 4, A and B). Anthocyanin biosynthesis is modulated by changes in Suc concentrations, and the sugar-dependent upregulation of the anthocyanin synthesis pathway is Suc-specific (Jeong et al. 2010). However, anthocyanin accumulation of ein3/eil1 leaves was higher than that of eil1-3 and ein3-1 (Figure 4, A and B), whereas glucose and sucrose levels of ein3/eil1 leaves were comparable with those of eil1-3 and ein3-1 (Figure 4, C–E). eil1-3 and ein3-1 are weak ethylene-insensitive mutants, and ein3/eil1 is a strongly insensitive mutant; sugar-induced anthocyanin accumulation is inhibited by ethylene in Arabidopsis (Jeong et al. 2010). Therefore, ethylene signaling was more severely interrupted in ein3/eil1 seedlings than in eil1-3 and ein3-1 seedlings. Although ein3-1, eil1-3, and ein3/eil1 mutants exhibited similar increases in glucose and sucrose, only ein3/eil1 mutants demonstrated increased anthocyanin levels. Collectively, the above findings indicated that the increase in anthocyanin accumulation in ein3-1, eil1-3, and ein3/eil1 mutants was related to endogenous sucrose accumulation.
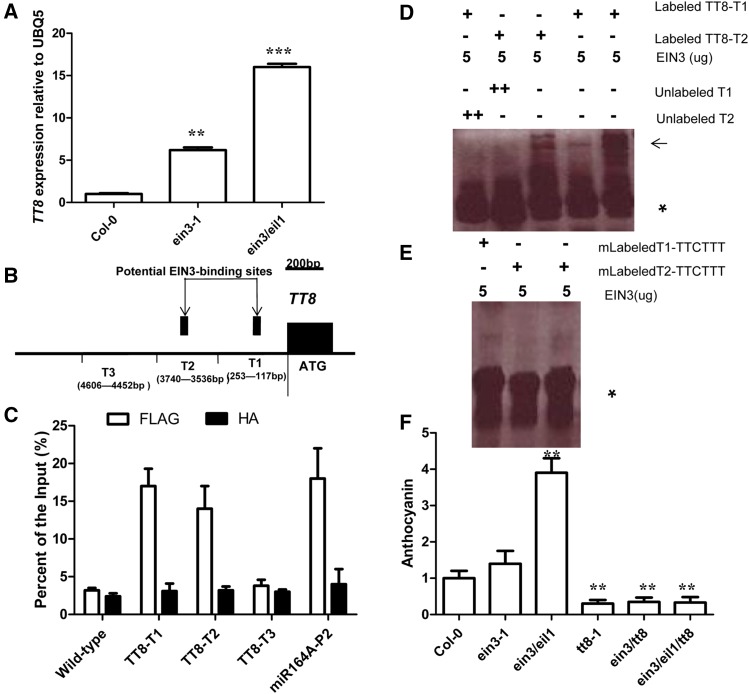
EIN3 negatively regulates TT8 via direct occupancy of its promoter
The above observations prompted us to examine whether EIN3 directly regulates the MBW transcription complex to control anthocyanin accumulation. Previous reports (Jeong et al. 2010) showed that expression levels of DFR/TT3, TT18, TT8, UF3GT, PAP2, and MYLB2 are dramatically upregulated in ein3-1 and ein3/eil1 mutants relative to wild-type (Col-0) seedlings. We confirmed this result in the case of TT8 by quantitative PCR (Figure 5A). We screened the promoters of these genes and found that their promoters’ sequences all contained a putative EIN3 binding site (EBS; TACAT or TTCAAA; Figure 5B; Li et al. 2013). We then determined potential EIN3 binding to these promoters by ChIP-PCR analysis. Our findings indicated that EIN3 bound to the EBSs of the T1 and T2 regions of the TT8 promoter (containing TTCAAA or TACAT sequences) and EBSs of the P2 region of the miR164A promoter when used as a positive control in vivo (Figure 5C; Li et al. 2013). By contrast, EIN3 could not bind to the EBSs of the T3 regions of the TT8 promoter and wild-type, which were used as negative control (Figure 5C). Although the promoter sequences of DFR/TT3, TT18, UF3GT, PAP2, and MYLB2 genes contained EBSs, EIN3 did not bind specifically to these promoter sequences (Figure S2). These results revealed that EIN3 did not bind to all EBSs. Thus, EIN3 binding to the promoter of TT8 in vivo might potentially require contributions from adjacent promoter sequences to support its binding specificity (Li et al. 2013). Additionally, EMSA experiments were performed to determine the in vitro binding of EIN3 to the TT8-T1 and TT8-2 EBS regions. Indeed, EIN3 could bind to the labeled TT8-T1 and TT8-2 elements in vitro (Figure 5D). Excessive unlabeled competitor DNA fragments effectively abolished this binding (Figure 5D). However, EIN3 was unable to bind mutant derivatives of EBSs from the TT8-T1 and TT8-2 regions of the cognate promoter (Figure 5E). Although the expression levels of PAP2 and MYBL2 were significantly enhanced in ein3/eil1 seedlings, those of TTG1, EGL3, and GL3 were not altered between ein3/eil1 and wild-type seedlings (Jeong et al. 2010; Figure S2, A and B). Therefore, we focused on PAP2 and MYBL2. However, our findings of ChIP-PCR indicated that EIN3 did not directly bind to the promoters of PAP2 and MYBL2 (Figure S2, C–F).
Figure 5.
TT8 is a target gene of EIN3. (A) Bar graph showing expression of TT8 between Col-0, ein3-1, and ein3/eil1 seedlings grown under white light conditions. Seedlings of at least five independently propagated lines were utilized. Wild-type data are set as 1.0. Quantification was normalized to the expression of UBQ5. Error bars represent SD (n = 4; ** P < 0.01, *** P < 0.001). (B) Schematic of the TT8 promoter loci and their amplicons for ChIP analysis. (C) Chromatin immunoprecipitation (ChIP) analysis. Enrichment of particular TT8 chromatin regions with anti-HA antibody (as a control) or anti-FLAG antibody in EIN3-FLAG transgenic plants and wild type as detected by real-time PCR analysis. Quantifications were normalized to the expression of UBQ5. Error bars represent SD (n = 4; ** P < 0.01). Input is set as 100% [supernatant including chromatin (input material) is considered as 100%, immunoprecipitated chromatin/input material × 100% for enrichment product of particular TT8 chromatin regions]. P2 promoter of miR164A is a positive control, and the primers used for the P2 promoter have been described previously (Li et al. 2013). (D) Unlabeled TT8 promoter (2.0 μg) was used as a competitor to determine the specificity of DNA-binding activity for EIN3. Free probe and EIN3 probe complexes are indicated by an asterisk and arrows, respectively. (E) A mutant version of the TT8 promoter (AAA/TTT) was labeled with biotin and used for EMSA with EIN3 polypeptides. Free probe is indicated by an asterisk. (F) Bar graph showing the difference in anthocyanin accumulation between all indicated developmental seedling (12-day-old) grown under white light on solid MS medium with 1% sucrose. Col-0 data set as 1.0. Error bars represent SD (n = 10; ** P < 0.01). Experiments were repeated two times with similar results.
The ein3/eil1 mutants showed higher anthocyanin accumulation than the control seedlings (12 days old), and the tt8-1 mutant exhibited significantly less anthocyanin accumulation in comparison (Nesi et al. 2000; Joeng et al. 2010; Figure 5F). Furthermore, ein3/tt8 and ein3/eil1/tt8 plants were found to have reduced anthocyanin accumulation relative to control seedlings (Figure 5F). Thus, loss of TT8 function suppressed anthocyanin accumulation in the ein3/eil1 double mutant. EIN3 acted genetically upstream of TT8, and EIN3/EIL1-TT8 formed a signaling cascade to regulate anthocyanin accumulation. Together, these findings indicated that TT8 was a target of EIN3.
Discussion
YDA-EIN3-TT8 controls anthocyanin accumulation via protein–protein interactions
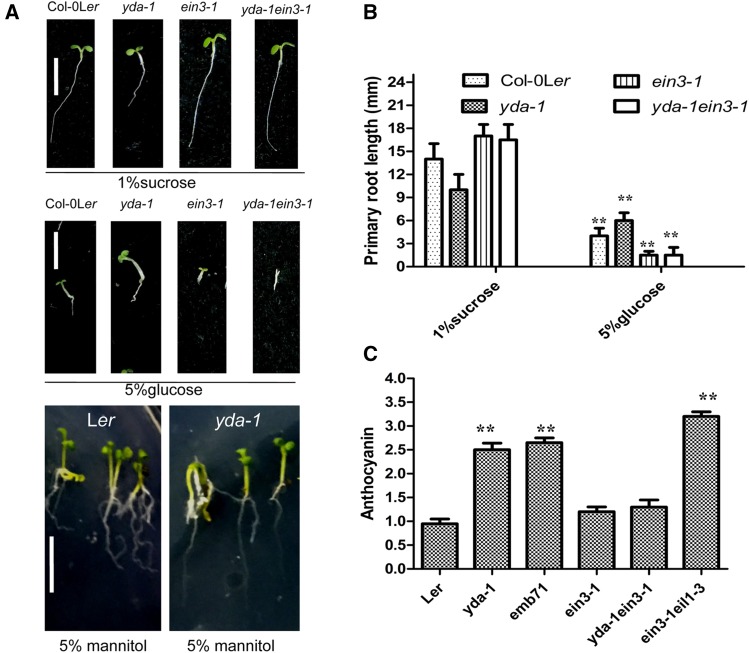
Although yda-1 and emb71 plants showed higher anthocyanin accumulation than control wild-type seedlings (Meng et al. 2016; Figure 6C), ein3-1 mutants presented comparable anthocyanin accumulation with wild-type seedlings (Figure 4 and Figure 6C; Joeng et al. 2010). Furthermore, yda/ein3 double-mutant seedlings presented comparable anthocyanin accumulation with wild-type and ein3-1 seedlings (Figure 6C), suggesting that EIN3 was downstream of YDA, and loss of EIN3 function significantly decreased anthocyanin content in yda-1 mutants. These findings suggested a close relationship between YDA and EIN3 during anthocyanin accumulation. YDA interacted with EIN3, as proven by in vitro or in vivo pull-down assays, Y2H, and BiFC (Figure 2 and Figure 3). Influence of the absence of YDA function on endogenous sugar content (Figure 4; Meng et al. 2016) and specific sugar sensing, but not osmotic stress in general (Figure 6, A–C; Yanagisawa et al. 2003; Meng et al. 2016), provided additional support for possible regulatory activities of YDA upon EIN3. Moreover, EMB71/YDA-EIN3 directly targeted TT8 by EIN3 (Figure 7), which may form an EIN3-TT8 gene cascade for the regulation of anthocyanin accumulation. TT8 is a key regulator of anthocyanin and proanthocyanidin biosynthesis in Arabidopsis (Xu et al. 2013). Collectively, these data suggested that a YDA interaction with the EIN3-TT8 complex might regulate anthocyanin accumulation.
Figure 6.
EIN3 mutation differentially suppresses yda mutant. (A) Representative 6-day-old seedlings grown on solid MS medium with 1% sucrose, 5% glucose, and 5% mannitol, and every kind of seedling have been indicated. Bar, 5 mm. (B) Bar graph exhibiting the difference in the root length between Col-0Ler, yda-1, ein3-1, and yda/ein3 seedlings grown on solid MS medium with 1% sucrose and 5% glucose, respectively. Error bars represent SD (n = 20; ** P < 0.01). Experiments were repeated two times with similar results. (C) Bar graph exhibiting the difference in the anthocyanin accumulation between the indicated seedlings grown under white light condition solid MS medium with 1% sucrose. Wild-type Ler is set as 1.0. Error bars represent SD (n = 13; ** P < 0.01). Experiments were repeated two times with similar results.
Figure 7.
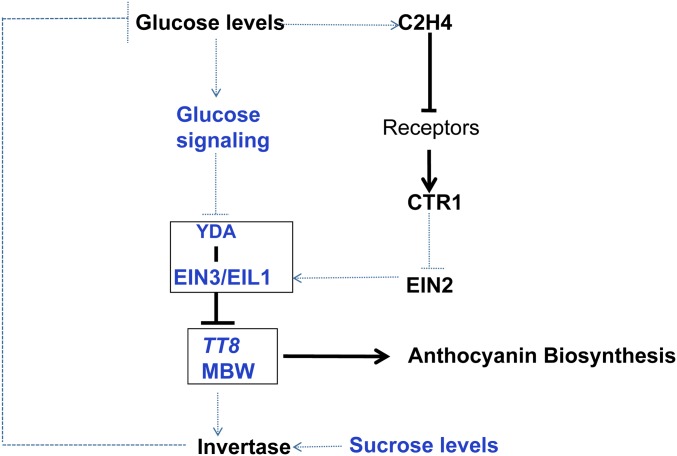
Model illustrating the regulation of anthocyanin biosynthesis by sugar and ethylene signaling. With exogenous sucrose supplied, increased sucrose levels may enhance invertase activity (Lou et al. 2007). Invertase can degrade sucrose into fructose and glucose, and generated glucose signaling/levels control YDA stability, which is due to glucose promoting YDA stability (Meng et al. 2016). And TT8, in turn, may control invertase activity by YDA-EIN3-TT8 signal transduction pathway (Payyavula et al. 2013). Thus, it appears to form a glucose signal loop for controlling sucrose-specifically depends on anthocyanin biosynthesis. This explains why anthocyanin biosynthesis specifically depends on sucrose. In a similar fashion, glucose signaling induces ethylene production, which also triggers the suppression of anthocyanin accumulation by the C2H4-Receptor-CTR1-EIN2-EIN3/EIL1 signal pathway. Solid lines indicate direct regulation, whereas dotted lines indicate either indirect regulation or regulation in an unknown manner.
The precise mechanism by which YDA regulates EIN3 is presently unknown, but we found that YDA directly interacted with EIN3, which is a common target of both ethylene and sugar signaling (Yanagisawa et al. 2003). EIN3 contains an amino acid sequence motif reminiscent of a MAP kinase substrate docking domain (Sharrocks et al. 2000; Andreasson et al. 2005), which suggested that YDA might interact with EIN3 at this site. Given that YDA belongs to the MEKK1/Ste11/Bck1 class of MAPKKKs, EIN3 stability/function might be affected following YDA interaction and subsequent phosphorylation. The two proposed EIN3 phosphorylation sites are thought to show dual functions: T174 for stabilization and T592 for degradation (Yoo et al. 2008), but the kinase(s) responsible for EIN3 phosphorylation remain to be rigorously determined (Yoo et al. 2008; An et al. 2010).
Anthocyanin accumulation in yda mutants is positively correlated with sucrose accumulation
The anthocyanin biosynthetic pathway is strongly activated in response to increasing Suc concentrations, and this sugar-dependent upregulation is Suc-specific (Solfanelli et al. 2006; Jeong et al. 2010). In this work, we suggested that the increased sucrose concentration in yda seedlings might be responsible for anthocyanin accumulation. Although plant hormones such as auxin, ABA (Jeong et al. 2004; Hoth et al. 2010), cytokinin (Deikman and Hammer 1995), gibberellins (Weiss et al. 1995), and ethylene (Morgan and Drew 1997; Jeong et al. 2010) differentially regulate anthocyanin biosynthesis (Ozeki and Komamine 1986), yda did not affect signaling by these hormones (Lukowitz et al. 2004). Therefore, increased anthocyanin accumulation in yda mutants might predominantly result from a high endogenous sucrose concentration (Meng et al. 2016; Figure 6).
Proposed model for the regulation of anthocyanin biosynthesis
Increased anthocyanin accumulation was observed in the ethylene-insensitive mutants etr1-1, ein2-1, and ein3/eil1. By contrast, the constitutive ethylene-response mutant ctr1-1, partial ethylene-insensitive mutant ein3-1, and weak ethylene-insensitive mutant eil1-3 exhibited comparable anthocyanin accumulation with wild-type seedlings (Jeong et al. 2010). These data were consistent with our results, which indicated that EIN3 and EIL1 were functionally redundant with respect to the regulation of anthocyanin accumulation. Thus, ethylene repressed the Suc-specific signaling pathway via the ethylene-mediated C2H4-Receptor-CTR1-EIN2-EIN3/EIL1-TT8 signaling cascade to regulate anthocyanin accumulation (Figure 7). Sugar and ethylene signaling were closely associated in the regulation of anthocyanin biosynthesis (Figure 7). Suc could enhance ethylene production in a dose-dependent manner in tobacco (Philosoph-Hadas et al. 1985), rice (Oryza sativa; Kobayashi and Saka 2000), and Arabidopsis (Jeong et al. 2010) leaves. Meanwhile, ethylene modulates Suc and Glc sensitivity during Arabidopsis seedling development (Gibson et al. 2001).
In numerous cases, sucrose is not directly the signaling molecule, and many experiments have proven the role of hexose in triggering sugar signaling and responses in plants (Sheen et al. 1999). The role of hexose as signaling molecule but not substrate was also confirmed through the observation that hexokinase is a dimeric cytosolic enzyme essential for glycolysis (Sheen et al. 1999). Invertase (INV) controls plant growth and provides glucose for sucrose-dependent anthocyanin biosynthesis by catalyzing sucrose catabolism, which is one of the largest metabolic fluxes in plants (Barratt et al. 2009). INV gene expression levels are enhanced when the PAP1-related potato MYB transcription factor Solanum tuberosum anthocyanin 1 (StAN1) is infiltrated into tobacco leaves (Payyavula et al. 2013). Therefore, the YDA-EIN3 complex directly and negatively regulates the promoter of TT8, thereby influencing the MBW complex. MBW might in turn regulate INV activity, which degrades sucrose into fructose and glucose (Lou et al. 2007). Increased glucose levels promote YDA stability (Meng et al. 2016), which mediates the YDA-EIN3-TT8 pathway to regulate anthocyanin biosynthesis (Figure 7).
Whether YDA-EIN3 functions independently of MBW components is currently unknown. However, this relationship should be determined because many abiotic stresses induce anthocyanin biosynthesis as part of a coordinated nonspecific response to stress. In this work, building on a previous report (Jeong et al. 2010), we found that TT8, a member of MBW, was a target of EIN3. On the basis of quantitative PCR, ChIP-PCR, EMSA, and genetic analyses, our findings suggested that YDA-EIN3-TT8 formed a sugar signaling-mediated gene cascade that regulated anthocyanin biosynthesis (Figure 7). Moreover, microarray analysis of pho3 plants, which are deficient in phosphorus and show high levels of anthocyanin accumulation, revealed an increased expression of TT8 and genes encoding anthocyanin biosynthetic enzymes, implying that sugar was a trigger of anthocyanin biosynthesis in vivo.
Collectively, our findings enabled the synthesis of a model for the function of the EMB71/YDA-EIN3-EIL1-TT8 and C2H4-Receptor-CTR1-EIN2-EIN3/EIL1-TT8 signaling cascades in the regulation of anthocyanin biosynthesis. Suc signaling induces anthocyanin accumulation by the EMB71/YDA-EIN3-EIL1-TT8 signaling cascade and simultaneously induces ethylene production (Jeong et al. 2010). Ethylene subsequently mediates the suppression of anthocyanin accumulation by the C2H4-Receptor-CTR1-EIN2-EIN3/EIL1-TT8 signaling cascade (Figure 7). Thus, an antagonistic interaction exists between the EMB71/YDA-EIN3-EIL1-TT8 and C2H4-Receptor-CTR1-EIN2-EIN3/EIL1-TT8 signaling cascades. EIN3 is a common target of both ethylene and sugar signaling (Yoo et al. 2008). EIN3/EIL1 is a central component that coordinates either sugar or ethylene signaling, with opposing consequences for anthocyanin biosynthesis.
Anthocyanins play a vital role in protecting plants against ultraviolet radiation, insect attack, and pathogen infection (Harborne and Williams 2000; Winkel-Shirley 2002; Gould 2004). However, the accumulation of anthocyanins above a threshold level is disadvantageous. For example, sustained anthocyanin accumulation can cause imbalances between primary and secondary metabolites (Jeong et al. 2010). Thus, plants may have evolved elaborate strategies to dynamically equilibrate anthocyanin biosynthesis within an effective but manageable range. Our findings suggested a molecular mechanism with multifaceted regulatory layers with which to finetune anthocyanin biosynthesis during plant growth and development.
Acknowledgments
We thank Gary. J. Loake (Edinburgh University) for kindly revising this manuscript. This work was funded by grants from the Natural Science Foundation of Jiangsu province of China (BK20170236) and the Postgraduate Research and Practice Innovation Program of Jiangsu Province (KYCX17-1614). The authors declare no competing financial interest.
Author contributions: L.-S.M. designed experiments. L.-S.M., M.-K.X., W.W., X.-Y.C., F.Y., Z.-Q.W., M.-J.L., C.L., and J.-Y.W. performed the experiments. L.-S.M. and M.-K.X. completed statistical analysis of data. L.-S.M., Z.-Y.L., and J.-H.J. wrote, edited, and revised this manuscript.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.6084/m9.figshare.6974570.
Communicating editor: N. Springer
Literature Cited
- An F., Zhao Q., Ji Y., Li W., Jiang Z., et al. , 2010. Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 22: 2384–2401. 10.1105/tpc.110.076588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasson E., Jenkins T., Brodersen P., Thorgrimsen S., Petersen N. H. T., et al. , 2005. The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J. 24: 2579–2589. 10.1038/sj.emboj.7600737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier M., Hemmann G., Holman R., Corke F., Card R., et al. , 2004. Characterization of mutants in Arabidopsis showing increased sugar-specific gene expression, growth, and developmental responses. Plant Physiol. 134: 81–91. 10.1104/pp.103.031674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt D. H. P., Derbyshire P., Findlay K., Pike M., Wellner N., et al. , 2009. Normal growth of Arabidopsis requires cytosolic invertase but not sucrose synthase. Proc. Natl. Acad. Sci. USA 106: 13124–13129. 10.1073/pnas.0900689106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudry A., Caboche M., Lepiniec L., 2006. TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 46: 768–779. 10.1111/j.1365-313X.2006.02733.x [DOI] [PubMed] [Google Scholar]
- Bemis S. M., Lee J. S., Shpak E. D., Torii K. U., 2013. Regulation of floral patterning and organ identity by Arabidopsis ERECTA-family receptor kinase genes. J. Exp. Bot. 64: 5323–5333. 10.1093/jxb/ert270 [DOI] [PubMed] [Google Scholar]
- Bergmann D. C., Lukowitz W., Somerville C. R., 2004. Stomatal development and pattern controlled by a MAPKK kinase. Science 304: 1494–1497. 10.1126/science.1096014 [DOI] [PubMed] [Google Scholar]
- Borevitz J. O., Xia Y., Blount J., Dixon R. A., Lamb C., 2000. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2394. 10.1105/tpc.12.12.2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Q., Rothenberg M., Solano R., Roman G., Terzaghi W., et al. , 1997. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89: 1133–1144. 10.1016/S0092-8674(00)80300-1 [DOI] [PubMed] [Google Scholar]
- Deikman J., Hammer P. E., 1995. Induction of anthocyanin accumulation by cytokinins in Arabidopsis thaliana. Plant Physiol. 108: 47–57. 10.1104/pp.108.1.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner H. K., Robbins T. P., Jorgensen R. A., 1991. Genetic and developmental control of anthocyanin biosynthesis. Annu. Rev. Genet. 25: 173–199. 10.1146/annurev.ge.25.120191.001133 [DOI] [PubMed] [Google Scholar]
- Gibson S. I., Laby R. J., Kim D., 2001. The sugar-insensitive1 (sis1) mutant of Arabidopsis is allelic to ctr1. Biochem. Biophys. Res. Commun. 280: 196–203. 10.1006/bbrc.2000.4062 [DOI] [PubMed] [Google Scholar]
- Gonzalez A., Zhao M., Leavitt J. M., Lloyd A. M., 2008. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/ Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53: 814–827. 10.1111/j.1365-313X.2007.03373.x [DOI] [PubMed] [Google Scholar]
- Gould K. S., 2004. Nature’s swiss army knife: the diverse protective roles of anthocyanins in leaves. J. Biomed. Biotechnol. 2004: 314–320. 10.1155/S1110724304406147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborne J. B., Williams C. A., 2000. Advances in flavonoid research since 1992. Phytochemistry 55: 481–504. 10.1016/S0031-9422(00)00235-1 [DOI] [PubMed] [Google Scholar]
- Hoth S., Niedermeier M., Feuerstein A., Hornig J., Sauer N., 2010. An ABA responsive element in the AtSUC1 promoter is involved in the regulation of AtSUC1 expression. Planta 232: 911–923. 10.1007/s00425-010-1228-4 [DOI] [PubMed] [Google Scholar]
- Jeong S. T., Goto-Yamamot N., Kobayashi S., Esaka M., 2004. Effects of plant hormones and shading on the accumulation of anthocyanins and the expression of anthocyanin biosynthetic genes in grape berry skins. Plant Sci. 167: 247–252. 10.1016/j.plantsci.2004.03.021 [DOI] [Google Scholar]
- Jeong S. W., Das P. K., Jeoung S. C., Song J. Y., Lee H. K., et al. , 2010. Ethylene suppression of sugar-induced anthocyanin pigmentation in Arabidopsis. Plant Physiol. 154: 1514–1531. 10.1104/pp.110.161869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C. Y., Lian H. L., Wang F. F., Huang J. R., Yang H. Q., 2009. Cryptochromes, phytochromes, and COP1 regulate light-controlled stomatal development in Arabidopsis. Plant Cell 21: 2624–2641. 10.1105/tpc.109.069765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H., Saka H., 2000. Relationship between ethylene evolution and sucrose content in excised leaf blades of rice. Plant Prod. Sci. 3: 398–403. 10.1626/pps.3.398 [DOI] [Google Scholar]
- Kubasek W. L., Shirley B. W., McKillop A., Goodman H. M., Briggs W., et al. , 1992. Regulation of flavonoid biosynthetic genes in germinating Arabidopsis seedlings. Plant Cell 4: 1229–1236. 10.1105/tpc.4.10.1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Ye C., Zhao R., Li X., Liu W.Z., et al. , 2015. Mitogen-activated protein kinase kinase kinase (MAPKKK) 4 from rapeseed (Brassica napus L.) is a novel member inducing ROS accumulation and cell death.. Biochem. Biophys. Res. Commun. 467: 792–797. 10.1016/j.bbrc.2015.10.063 [DOI] [PubMed] [Google Scholar]
- Li J., Wen J., Lease K. A., Doke J. T., Taxl F. E., et al. , 2002. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI and modulates brassinosteroid signaling. Cell 110: 213–222. 10.1016/S0092-8674(02)00812-7 [DOI] [PubMed] [Google Scholar]
- Li Z., Peng J. Y., Wen X., Guo H. W., 2013. ETHYLENE-INSENSITIVE3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 25: 3311–3328. 10.1105/tpc.113.113340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd J. C., Zakhleniuk O. V., 2004. Responses of primary and secondary metabolism to sugar accumulation revealed by microarray expression analysis of the Arabidopsis mutant, pho3. J. Exp. Bot. 55: 1221–1230. 10.1093/jxb/erh143 [DOI] [PubMed] [Google Scholar]
- Lou Y., Gou J. Y., Xue H. W., 2007. PIP5K9, an Arabidopsis phosphatidylinositol monophosphate kinase, interacts with a cytosolic invertase to negatively regulate sugar-mediated root growth. Plant Cell 19: 163–181. 10.1105/tpc.106.045658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W., Roeder A., Parmentr D., Somerville C., 2004. A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell 116: 109–119. 10.1016/S0092-8674(03)01067-5 [DOI] [PubMed] [Google Scholar]
- Meng L. S., 2015. Transcription coactivator Arabidopsis ANGUSTIFOLIA3 modulates anthocyanin accumulation and light-induced root elongation through transrepression of constitutive Photomorphogenic1. Plant Cell Environ. 38: 838–851. 10.1111/pce.12456 [DOI] [PubMed] [Google Scholar]
- Meng L. S., Yao S. Q., 2015. Transcription co-activator Arabidopsis ANGUSTIFOLIA3 (AN3) regulates water-use efficiency and drought tolerance by modulating stomatal density and improving root architecture by the transrepression of YODA (YDA). Plant Biotechnol. J. 13: 893–902. 10.1111/pbi.12324 [DOI] [PubMed] [Google Scholar]
- Meng L. S., Wang Y. B., Yao S. Q., Liu A., 2015a Arabidopsis AINTEGUMENTA (ANT) mediates salt tolerance by trans-repressing SCABP8. J. Cell Sci. 128: 2919–2927. 10.1242/jcs.172072 [DOI] [PubMed] [Google Scholar]
- Meng L. S., Wang Z. B., Yao S. Q., Liu A., 2015b The ARF2–ANT–COR15A gene cascade regulates ABA signaling-mediated resistance of large seeds to drought in Arabidopsis. J. Cell Sci. 128: 3922–3932. 10.1242/jcs.171207 [DOI] [PubMed] [Google Scholar]
- Meng L. S., Li Y. Q., Liu M. Q., Jiang J. H., 2016. The Arabidopsis ANGUSTIFOLIA3-YODA gene cascade induces anthocyanin accumulation by regulating sucrose levels. Front. Plant Sci. 7: 1728 [corrigenda: Front. Plant Sci. 8: 1228 (2017)] 10.3389/fpls.2016.01728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L. S., Li C., Xu M. K., SUN X. D., Wan W., et al. , 2018. Arabidopsis ANGUSTIFOLIA3 (AN3) is associated with the promoter of CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) to regulate light‐mediated stomatal development. Plant Cell Environ. 41: 1645–1656. 10.1111/pce.13212 [DOI] [PubMed] [Google Scholar]
- Morgan P. W., Drew M. C., 1997. Ethylene and plant responses to stress. Physiol. Plant. 100: 620–630. 10.1111/j.1399-3054.1997.tb03068.x [DOI] [Google Scholar]
- Nesi N., Debeaujon I., Jond C., Pelletier G., Caboche M., et al. , 2000. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12: 1863–1878. 10.1105/tpc.12.10.1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozeki Y., Komamine A., 1986. Effects of growth regulators on the induction of anthocyanin synthesis in carrot suspension cultures. Plant Cell Environ. 27: 1361–1368. 10.1093/oxfordjournals.pcp.a077234 [DOI] [Google Scholar]
- Payne C. T., Zhang F., Lloyd A. M., 2000. GL3 encodes a bHLH protein that regulates trichome development in arabidopsis through interaction with GL1 and TTG1. Genetics 156: 1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payyavula R. S., Singh R. K., Navarre D. A., 2013. Transcription factors, sucrose, and sucrose metabolic genes interact to regulate potato phenylpropanoid metabolism. J. Exp. Bot. 64: 5115–5131. 10.1093/jxb/ert303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philosoph-Hadas S., Meir S., Aharoni N., 1985. Carbohydrates stimulate ethylene production in tobacco leaf discs. II. Sites of stimulation in the ethylene biosynthesis pathway. Plant Physiol. 78: 139–143. 10.1104/pp.78.1.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T. C., Song S. S., Ren Q. C., Wu D. W., Huang H., et al. , 2011. The jasmonate-ZIM-domain proteins interact with the WD-repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23: 1795–1814. 10.1105/tpc.111.083261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabino I., Mancinelli A. L., 1986. Light, temperature, and anthocyanin production. Plant Physiol. 81: 922–924. 10.1104/pp.81.3.922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan D. D., Cao M., Lin-Wang K., Cooney J. M., Jensen D. J., et al. , 2009. Environmental regulation of leaf colour in red 35S:PAP1 Arabidopsis thaliana. New Phytol. 182: 102–115. 10.1111/j.1469-8137.2008.02737.x [DOI] [PubMed] [Google Scholar]
- Sharrocks A. D., Yang S. H., Galanis A., 2000. Docking domains and substrate-specificity determination for MAP kinases. Trends Biochem. Sci. 25: 448–453. 10.1016/S0968-0004(00)01627-3 [DOI] [PubMed] [Google Scholar]
- Sheen J., Zhou L., Jang J. C., 1999. Sugars as signaling molecules. Curr. Opin. Plant Biol. 2: 410–418. [DOI] [PubMed] [Google Scholar]
- Shirley B. W., Kubasek W. L., Storz G., Bruggemann E., Koornneef M., et al. , 1995. Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 8: 659–671. 10.1046/j.1365-313X.1995.08050659.x [DOI] [PubMed] [Google Scholar]
- Solfanelli C., Poggi A., Loreti E., Alpi A., Perata P., 2006. Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol. 140: 637–646. 10.1104/pp.105.072579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R., Werber M., Weisshaar B., 2001. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4: 447–456. 10.1016/S1369-5266(00)00199-0 [DOI] [PubMed] [Google Scholar]
- Stracke R., Ishihara H., Huep G., Barsch A., Mehrtens F., et al. , 2007. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 50: 660–677. 10.1111/j.1365-313X.2007.03078.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S., Keurentjes J., Bentsink L., Koornneef M., Smeekens S., 2005. Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 139: 1840–1852. 10.1104/pp.105.066688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E., Quail P. H., 2003. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770. 10.1105/tpc.013839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker A. R., Davison P. A., Bolognesi-Winfield A. C., James C. M., Srinivasan N., et al. , 1999. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11: 1337–1350. 10.1105/tpc.11.7.1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss D., Van Der Luit A., Knegt E., Vermeer E., Mol J., et al. , 1995. Identification of endogenous gibberellins in petunia flowers (induction of anthocyanin biosynthetic gene expression and the antagonistic effect of abscisic acid). Plant Physiol. 107: 695–702. 10.1104/pp.107.3.695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B., 2001. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126: 485–493. 10.1104/pp.126.2.485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B., 2002. Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 5: 218–223. 10.1016/S1369-5266(02)00256-X [DOI] [PubMed] [Google Scholar]
- Xu W., Grain D., Le Gourrierec J., Harscoët E., Berger A., et al. , 2013. Regulation of flavonoid biosynthesis involves an unexpected complex transcriptional regulation of TT8 expression, in Arabidopsis. New Phytol. 198: 59–70. 10.1111/nph.12142 [DOI] [PubMed] [Google Scholar]
- Yanagisawa S., Yoo S. D., Sheen J., 2003. Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 425: 521–525. 10.1038/nature01984 [DOI] [PubMed] [Google Scholar]
- Yoo S. D., Cho Y. H., Tena G., Xiong Y., Sheen J., 2008. Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451: 789–795. 10.1038/nature06543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Gonzalez A., Zhao M., Payne C. T., Lloyd A., 2003. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869. 10.1242/dev.00681 [DOI] [PubMed] [Google Scholar]
- Zimmermann I. M., Heim M. A., Weisshaar B., Uhrig J. F., 2004. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 40: 22–34. 10.1111/j.1365-313X.2004.02183.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors affirm that all data necessary for confirming the conclusions of the article are present within the article and figures. Strains and/or plasmids are available upon request. Supplemental figures are available in supporting figures and legends file. Supplemental Material, File S1 shows that HXK1, SUC1, and CINV2 cannot directly interacts with YDA in vitro. File S2 shows that PAP2 and MYBL2 are not target genes of EIN3. All genomic data and genomic sequencing raw reads are available online at https://www.arabidopsis.org/. Supplemental material available at Figshare: https://doi.org/10.6084/m9.figshare.6974570.