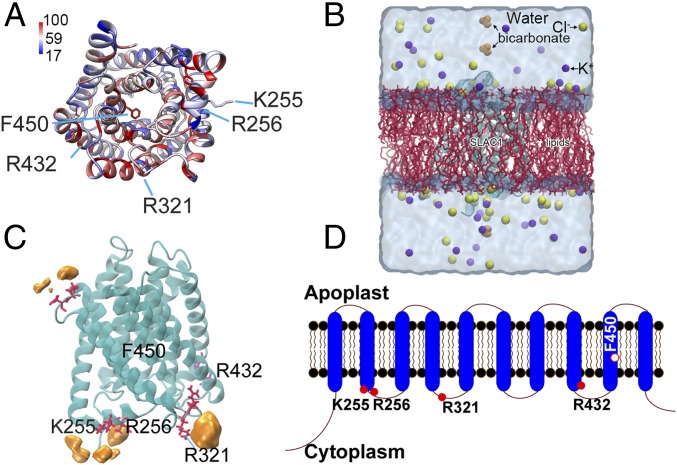
Fig. 1.
Computationally predicted SLAC1 protein structure and its potential bicarbonate binding sites. (A) Predicted structure of SLAC1 transmembrane domains. The structure was predicted as described using modeling algorithms and coordinates of the TehA template structure (35) and is pseudocolored based on sequence conservation among Arabidopsis SLAC/SLAH homologs and TehA. Blue and red respectively correspond to 17% and 100% conservation with AtSLAC1/SLAH homologous and TehA. (B) The simulation system is shown. The SLAC1 structure was embedded in a POPC lipid membrane using the CHARMM-GUI Membrane Builder. Bicarbonate was added to the model afterward (see Materials and Methods). The final simulation system of SLAC1 had dimensions of 83 × 83 × 105 Å3, with a total of ∼65,000 atoms. (C) Key residues (red) and probability of bicarbonate interaction map (orange). F450 is the proposed gate residue of SLAC1; K255, R256, and R321 are residues predicted to interact with bicarbonate during GaMD simulations; and R432 is the conserved control residue for which bicarbonate is predicted not to bind in simulations. (D) The SLAC1 protein is predicted to form 10 transmembrane α-helices with the N and C termini located in the cytoplasm. Filled red circles indicate the location of amino acids shown in A and C; the open red circle indicates the proposed channel gate residue.