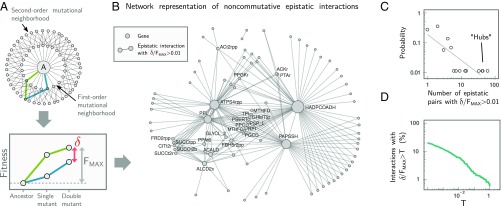
Fig. 3.
Short-range deformability in E. coli is rare, weak, and directional. (A) Systematic exploration of the second-order mutational neighborhood of E. coli. We exhaustively simulated every possible mutational trajectory starting from the ancestral (A) metabolism and ending in each double mutant. Noncommutative epistasis (δ) was measured for each pair of mutants and was normalized by FMAX, i.e., the maximal cumulative fitness effect of the double mutant: FMAX = max[ |Fi(A) + Fij(i)|, |Fj(A) + Fij(j)| ], where, e.g., Fx(y) denotes the fitness of mutant x when invading its immediate ancestor y at low frequency (Materials and Methods). (B) Network representation of all noncommutative epistatic pairs. Nodes represent mutations, and two nodes are joined by an edge if δ/FMAX >0.01 for that pair. Node labels (BiGG database notation) are shown for hubs (mutations with more than four noncommutative interactions). (C) Distribution of deformability for each gene in the network, measured as the number of other genes with which it has a noncommutative epistatic interaction. (D) Strength of all noncommutative epistatic interactions, i.e., the percentage of epistatic pairs with δ/FMAX > T as a function of T.