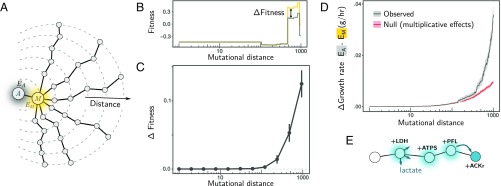
Fig. 4.
Long-range deformability of the E. coli metabolic fitness landscape. (A) We performed random walks (length = 1,000 mutations) in genotype space starting from an E. coli ancestor (A; gray) and first passing through a mutant (M; orange) with large environmental effect. (B and C) Fitness of mutants along these random walks was measured in competition with A (gray) in the environment it generates (EA), as well as in competition with M (orange) in the environment it generates (EM). In B we show the result for a single example of a random walk. Note that fitness in competition with M is shifted by the difference in fitness between M and A so all observed differences in fitness are due to deformation (for any genotype G, ΔFitness = |FG(A) − FG(M) − FM(A) |). (C) Average ΔFitness at increasing mutational distances from A in over n = 100 random walks (error bars represent SEM; n = 100). (D) Average difference (absolute value) in growth rate between environments EM and EA (in grams of dry cell weight × h−1) at varying genotype distances (gray line; shading represents SEM; n = 1,000). In red, we show the predicted difference in growth rates for a null model that assumes independent effects of mutations (Materials and Methods). (E) An example of an adaptive trajectory showing complex genetic interactions in a long-range deformation. The addition of LDH leads to the release of lactate as a by-product. Three additional mutations, ACKr, ATPS, and PFL, are required together for lactate to be used by a descendant genotype.