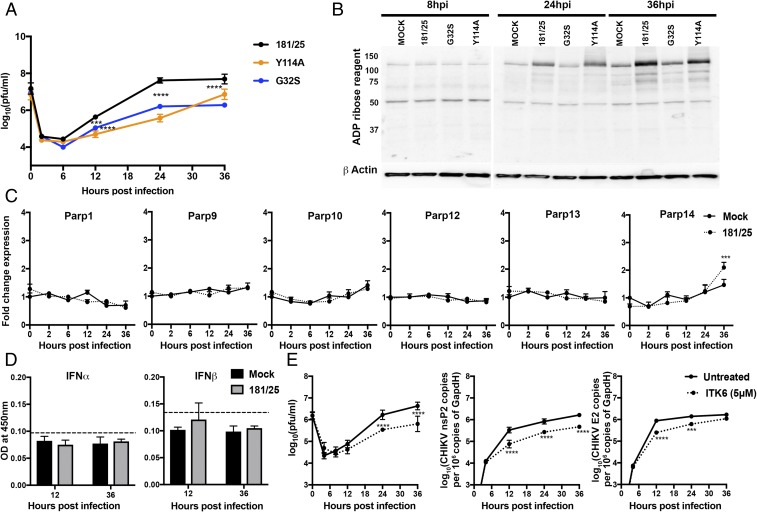
Fig. 1.
CHIKV infection facilitates virus replication in NSC34 cells by increasing ADP ribosylation of cellular proteins without inducing PARP gene transcription. (A) NSC34 cells were infected with CHIKV 181/25 (WT) and nsP3MD mutants G32S and Y114A at an MOI of 10. Virus production was measured by plaque formation in Vero cells. Each value represents the average from three independent experiments; error bars indicate SDs. ***P < 0.001, ****P < 0.0001 (181/25 vs. nsP3MD mutants G32S and Y114A). (Adapted with permission from ref. 27.) (B) NSC34 cells were infected with CHIKV 181/25 and nsP3MD mutants G32S and Y114A at an MOI of 5, and cell lysates were immunoblotted for ADPr. Antibody against β-actin was used for loading controls. A representative image from three independent experiments is shown. (C) NSC34 cells were infected with CHIKV 181/25 at an MOI of 5, and PARP mRNA expression was measured by qRT-PCR. Mock-infected cells (solid line) were compared with CHIKV-infected cells (dashed line). CT values were normalized to Gapdh, and fold change was calculated relative to uninfected 0-h (ΔΔCT) data. Each value represents the average ± SD from three independent experiments. ***P < 0.001 (mock vs. infected). (D) NSC34 cells were infected with CHIKV 181/25 (MOI = 5), and supernatant fluids were assayed for IFNα and IFNβ by enzyme immunoassay. Dashed lines indicate the lower limit of detection for the assays. (E) NSC34 cells were infected with CHIKV 181/25 (MOI = 1) and were treated or not treated with the pan mono-ADPr inhibitor ITK6 (5 μM) at the time of infection. Supernatant fluids were assayed for virus production by plaque formation in Vero cells (Left), and intracellular RNAs were assayed by qRT-PCR for viral genomic (nsP2; Middle) and genomic+sg E2 RNA and were compared with standard curves of CHIKV RNAs (Right). Each value represents the average ± SD from three independent experiments. ***P < 0.001, ****P < 0.0001.