Significance
Familial hypertrophic cardiomyopathy is a major cause of sudden death in young athletes. The mouse model for heart disease showed that the severe R403Q mutation in α-cardiac myosin heavy chain leads to increased ATPase activity and in vitro motility. But the β-myosin heavy chain is the preferred isoform in all larger mammals, leading us to generate a transgenic R403Q rabbit model. In contrast to the mouse model, we find a significant reduction in actin motility by R403Q rabbit myosin. Even more importantly, single myofibrils with the mutant myosin in the environment of a native thick filament showed diminished kinetics, force, and power output, leading to the conclusion that loss-of-function is the initial cause of cardiac hypertrophy.
Keywords: hypertrophic cardiomyopathy, cardiac myosin, myofibril kinetics, myofibril power output
Abstract
In 1990, the Seidmans showed that a single point mutation, R403Q, in the human β-myosin heavy chain (MHC) of heart muscle caused a particularly malignant form of familial hypertrophic cardiomyopathy (HCM) [Geisterfer-Lowrance AA, et al. (1990) Cell 62:999–1006.]. Since then, more than 300 mutations in the β-MHC have been reported, and yet there remains a poor understanding of how a single missense mutation in the MYH7 gene can lead to heart disease. Previous studies with a transgenic mouse model showed that the myosin phenotype depended on whether the mutation was in an α- or β-MHC backbone. This led to the generation of a transgenic rabbit model with the R403Q mutation in a β-MHC backbone. We find that the in vitro motility of heterodimeric R403Q myosin is markedly reduced, whereas the actin-activated ATPase activity of R403Q subfragment-1 is about the same as myosin from a nontransgenic littermate. Single myofibrils isolated from the ventricles of R403Q transgenic rabbits and analyzed by atomic force microscopy showed reduced rates of force development and relaxation, and achieved a significantly lower steady-state level of isometric force compared with nontransgenic myofibrils. Myofibrils isolated from the soleus gave similar results. The force–velocity relationship determined for R403Q ventricular myofibrils showed a decrease in the velocity of shortening under load, resulting in a diminished power output. We conclude that independent of whether experiments are performed with isolated molecules or with ordered molecules in the native thick filament of a myofibril, there is a loss-of-function induced by the R403Q mutation in β-cardiac myosin.
Since the seminal discovery by the Seidmans in 1990 (1) that a point mutation of arginine to glutamine (R403Q) in the human β-myosin heavy chain (MHC) caused a particularly malignant form of inherited hypertrophic cardiomyopathy (HCM), more than 300 mutations in the β-MHC have been reported that lead to heart disease. Several expression systems have been used to better understand the mechanism by which a single missense mutation in the MYH7 gene encoding the β-MHC can trigger a diseased state (reviewed in ref. 2). Due to the high degree of sequence and structural homology among all class II myosins, it was widely assumed that an HCM mutation would have the same effect on function independent of whether the myosin was extracted from human ventricular or soleus muscle (3–5), expressed as mutant smooth muscle myosin in the baculovirus/Sf9 insect cell system (6), or expressed as mutant Dictyostelium discoideum myosin in D. discoideum myosin-null cells (7). Unfortunately, a striated muscle myosin, such as cardiac myosin, could not be reproducibly expressed in Sf9 insect cells in a stable and active form (8, 9). Recently, a short subfragment-1 (S1) of human β-MHC has been expressed in a mouse myogenic cell line; this approach has made it possible to examine the functional changes introduced by mutations in human cardiac myosin (10–13), but the small magnitude and variability of the induced changes have not led to a unifying mechanism.
With the development of the mouse model for R403Q in 1996 (14), the mouse became a favorite organism for studies of HCM, given the ease of genetic manipulation and the rapid onset of many of the cardiac phenotypes characteristic of human disease (reviewed in ref. 15). Surprisingly, myosin prepared from mouse hearts homozygous for R403Q showed enhanced actin-activated ATPase activity and increased in vitro actin motility (16), suggesting a “gain-of-function” in contrast to the earlier results with human cardiac myosin that showed a “loss-of-function” (3, 4). Moreover, cardiac fibers from mice heterozygous for R403Q showed enhanced muscle mechanics consistent with the protein findings (17). Other mutations at myosin sites implicated in HCM also tended to show a gain-of-function in the mouse model (15).
A long recognized limitation of these studies is that the adult mouse expresses only α-MHC in its heart, whereas all larger mammals express predominantly β-MHC in their ventricles. This difference in isoform composition reflects the need of a small animal to optimize its power requirements with the faster α-MHC cardiac isoform (18, 19). Because α- and β-MHC are 93% identical in sequence, it was assumed that mutations introduced by gene-targeting or transgenesis would have a similar phenotype regardless of isoform composition. However, when the R403Q mutation was introduced into the mouse in either an α- or a β-MHC backbone by transgenesis (20), the results showed a markedly different phenotype depending on the myosin isoform: the R403Q mutation produced a significant enhancement in actin-activated, steady-state ATPase activity and transient kinetics in an α-MHC (S1) backbone, but not in a β-MHC (S1) backbone, where no change or a slight loss in activity was observed (20, 21). The low level of transgenic R403Q β-MHC expression, and the predominant endogenous α-MHC background in the mouse, precluded a more extensive investigation at the molecular and fiber levels. Thus, despite more than two decades of research into the causal effect of cardiac myosin mutations, the gain- vs. loss-of-function remains unresolved.
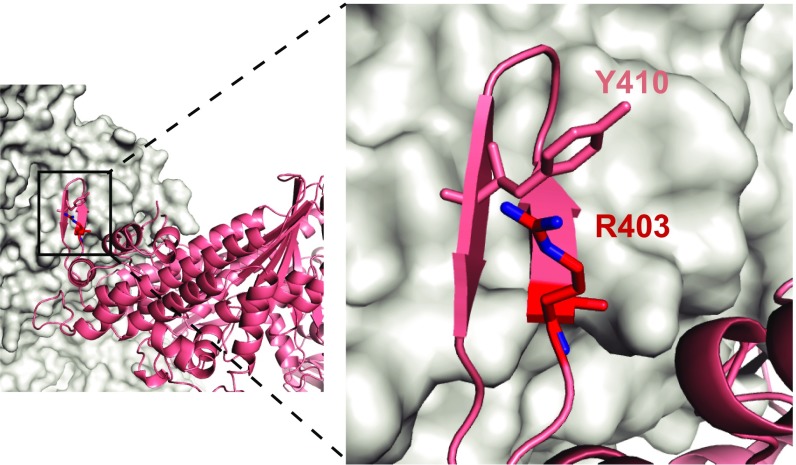
The need for a large animal model that expresses predominantly β-MHC in its ventricles was fulfilled by the generation of a transgenic (TG) rabbit carrying the R403Q mutation in a human β-MHC backbone that fully recapitulated the disease phenotype of HCM (22). Further insight into the pathogenesis of HCM was obtained by following the development of cardiac phenotypes in a large cohort of TG R403Q rabbits and their nontransgenic (NTG) littermates over a period of 4 y (23). Unfortunately, no mechanistic studies were undertaken at the time and this line expired. We have created a similar transgenic rabbit, but the R403Q mutation was introduced into a full-length rabbit cDNA construct to preserve a homologous system in which both the mutant and endogenous myosin have a rabbit β-MHC and rabbit light chains. A rabbit β-MHC promoter was used to drive expression of β-myosin in the soleus as well as in the ventricles (24). Here we present a description of the functional effects of the R403Q mutation in rabbit β-cardiac myosin at the molecular and myofibrillar levels. Given the location of the R403Q mutation at the base of a surface loop that interacts directly with actin (Fig. 1), it is not too surprising that a loss-of-function was observed in both myosin molecules and isolated myofibrils, leading to a decrease in the power output of the heart.
Fig. 1.
Model of the cardiomyopathy loop at the actin–myosin interface. Cryoelectron microscopy was used to obtain the structure of a human nonmuscle myosin bound to Factin/tropomyosin at near-atomic resolution, 3.9 Å (43). The CM-loop (salmon) forms an antiparallel β-strand that is the major site of interaction of the upper 50-kDa motor domain with actin (gray). This highly ordered loop is conserved in all myosin II isoforms and is stabilized primarily by hydrophobic interactions with actin. A model for the effect of the R403Q mutation in β-cardiac myosin proposes that arginine 403 (red) interacts with a tyrosine residue on the neighboring β-strand (PDB ID code 5JLH), and a mutation at this site weakens the interaction of the CM-loop with actin (43).
Results
TG rabbits carrying the R403Q mutation showed no increase in morbidity or mortality over a 2-y period. The primary functional effects of the mutation in muscle tissue could thus be more readily distinguished from secondary effects due to cardiac remodeling. Importantly, unlike the mouse model, we could isolate heterodimers of full-length myosin with one mutant R403Q heavy chain bound to a native β-MHC, given the endogenous β-MHC composition of the rabbit ventricles. This made it possible to do in vitro motility assays with intact myosin, which showed a consistent decrease in actin filament velocity by the mutant β-cardiac myosin compared with NTG controls. Similarly, given the β-MHC background, it was possible to show a loss in force, kinetic parameters, and power output in single mutant myofibrils. The preparation of myofibrils avoids the pathological hallmark of HCM, which is myocyte disarray in addition to hypertrophy and fibrosis.
Isolation and Quantification of R403Q Cardiac β-Myosin.
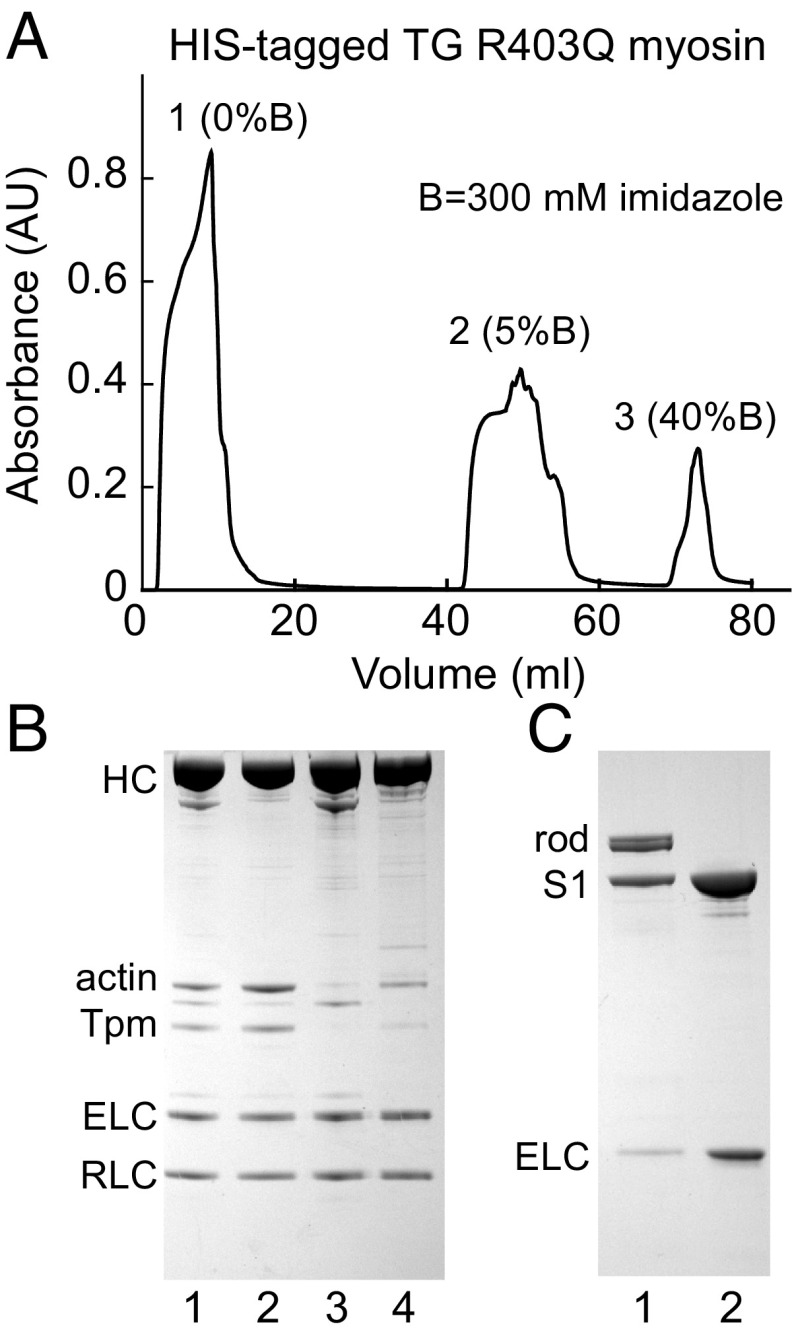
Transgenic His-tagged mutant rabbit myosin was isolated by nickel-chelating affinity chromatography, as described in detail for the TG mouse (20). In brief, crude myosin extracted from 1 to 2 g of homogenized heart tissue was clarified and loaded onto a Ni2+-chelating column equilibrated in 0.5 M NaCl, 20 mM Hepes, pH 7.5, and eluted with an imidazole gradient (Fig. 2A). The quality of the purified His-tagged R403Q rabbit cardiac myosin was very good as judged by SDS gel electrophoresis (Fig. 2B). SDS/glycerol separation gels gave no indication of any α-myosin in the crude or purified TG rabbit myosin. This is to be expected from earlier immunological analysis of peptides, which showed that α-myosin is only 10–20% of rabbit ventricular myosin by 3 mo of age, and our hearts were >8 mo to 1-y-old (25). From the final concentration of purified myosin (∼10%) relative to total myosin applied to a nickel-affinity column (Fig. 2A), and the level of mutant RNA (∼33%) expressed in the TG rabbit relative to wild-type (SI Appendix, SI Materials and Methods), we conclude that a minimum of 10% of two-headed myosin molecules contain a mutated motor domain. It has always been assumed that a low concentration of heavy chain with a single missense mutation would associate randomly with the native heavy chain to form a heterodimer (3). Moreover, in our previous paper (20) on His-tagged mouse R403Q β-myosin expressed in the native α-myosin mouse, we could show directly on SDS/glycerol gels that nickel-affinity–purified myosin had equal concentrations of α- and β-myosin heavy chain, which were well separated on these gels.
Fig. 2.
(A) His-tagged R403Q rabbit myosin was applied to a nickel-chelating column: peak 1 was the flow-through; peak 2 contained nonspecifically bound myosin eluted at 15 mM imidazole; and peak 3 was specifically bound His-tagged myosin eluted at 120 mM imidazole. Anti-His antibody reacted with peaks 1 and 3, but not with peak 2. Any reactive myosin must contain the R403Q mutation on at least one chain because R403Q was cloned together with the His-tag on the same cDNA construct. The percent mutant protein estimated from the yield of peak 3 divided by the total protein yield from the three peaks is about 10% R403Q myosin. (B) SDS gel of pooled fractions: (lane 1) initial crude myosin, (lane 2) flow-through fraction, (lane 3) nonspecifically bound fraction, and (lane 4) specifically bound myosin eluted at 120 mM imidazole. (C) Crude His-tagged myosin was digested with α-chymotrypsin at low ionic strength, and the supernatant (lane 1) was applied to the same chelating column as used for myosin, with pure R403Q S1 eluted at 120 mM imidazole (lane 2).
The Actin-Activated ATPase Activity of R403Q Rabbit Cardiac S1 Is Similar to That of NTG Rabbit Controls.
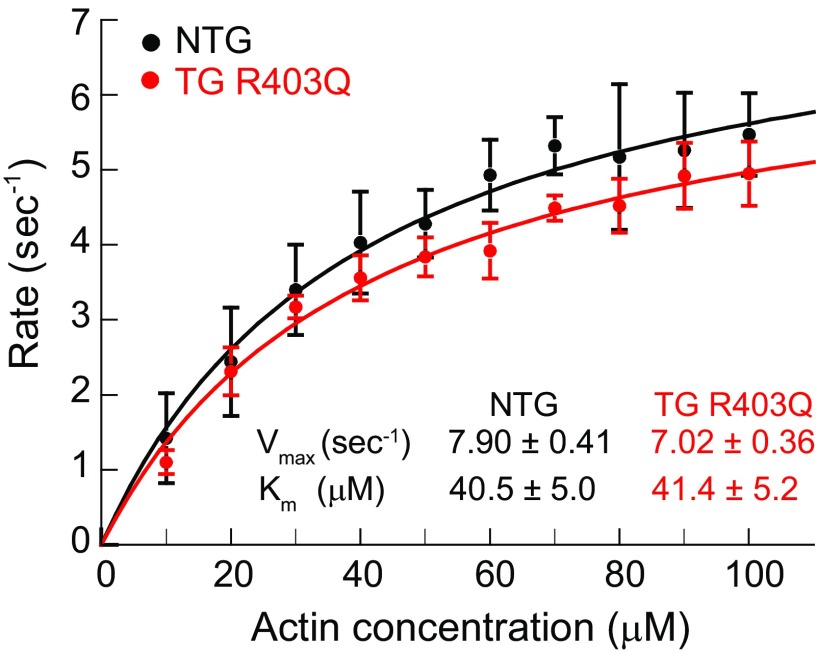
His6-tagged mutant S1 was prepared from R403Q rabbit myosin by chymotryptic digestion, and subsequent purification by nickel-chelating affinity chromatography (20). Untagged nontransgenic S1 was isolated from NTG rabbit myosin by the same proteolytic digestion, but purified by ion-exchange chromatography (26). SDS gels showed a single heavy chain and essential light chain (ELC) (Fig. 2C), characteristic of S1s isolated from striated muscle class II myosin (20, 21); the regulatory light chain (RLC) is degraded by the enzyme. The actin-activated ATPase activity from multiple R403Q preparations compared with NTG controls is shown in Fig. 3. Although some rabbit heart preparations indicated a slightly lower maximal velocity (Vmax) for R403Q S1, the majority of the kinetic assays showed no significant difference (P = 0.065) between the mutant S1 and control (SI Appendix, Table S1).
Fig. 3.
Actin-activated MgATPase activity for mutant R403Q rabbit S1 and NTG rabbit S1 at 30 °C. The data represent an average of multiple measurements from at least three independent S1 preparations. Error bars are SD. The data were fit to the Michaelis–Menten equation to obtain Vmax and Km values. NTG S1 has a Vmax of 7.90 ± 0.41 s−1 (95% CI 7.09–8.94) compared with 7.02 ± 0.36 s−1 (95% CI 6.30–7.98) for R403Q S1. The Km values were also similar at ∼41 µM.
The in Vitro Motility of R403Q Rabbit Myosin Is Markedly Reduced Relative to NTG Controls.
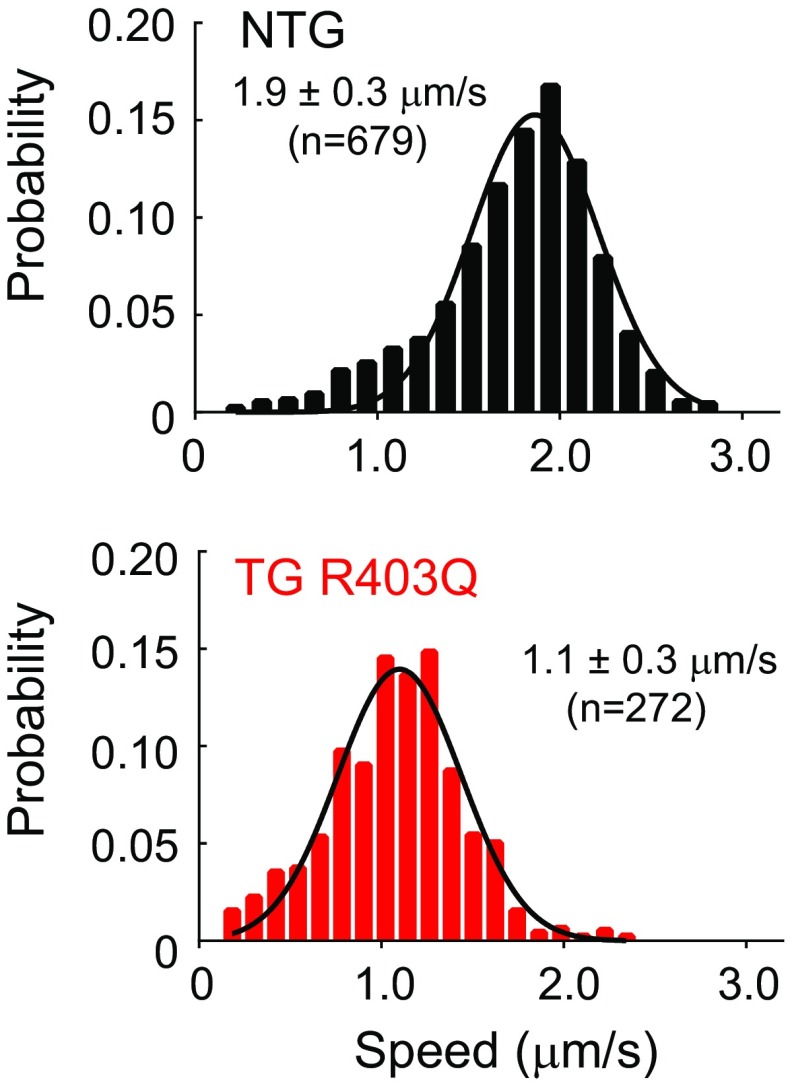
Analysis of the movement of several hundred actin filaments by purified myosin adhered nonspecifically to a nitrocellulose coated coverslip showed a markedly lower velocity of ∼1 µm/s for heterodimeric R403Q myosin compared with values of ∼2 µm/s for NTG myosin (Fig. 4). This twofold difference in motility may be an overestimation, because NTG myosin purified by hydrophobic interaction chromatography (HIC) selects for the most native, homogeneous myosin population, which can lead to a slightly higher velocity value than myosin purified by more conventional chromatographic methods (19). However, this concern is minimal, as the His6-tagged wild-type mouse α- or β-myosin purified by nickel-affinity chromatography showed the same motility as untagged mouse cardiac isoforms purified by HIC (20). Considering that the R403Q myosin has primarily only one of its two heavy chains mutated, the consistently lower motility suggests a significant impairment in the actin–myosin interaction.
Fig. 4.
Actin filament velocities for R403Q myosin compared with native rabbit myosin by in vitro motility assays at 30 °C. (Upper) Histogram is from NTG myosin; (Lower) histogramfrom TG myosin. The number of moving filaments is n. Each histogram is fit with a Gaussian to obtain the mean velocity and SD (20). The two means are significantly different (P < 0.0001, z test). At least three independent preparations gave mean velocities similar to the representative dataset shown here.
The Force and Kinetics of Attachment and Detachment of Myosin to Thin Filaments Are Reduced in Single Rabbit R403Q Myofibrils.
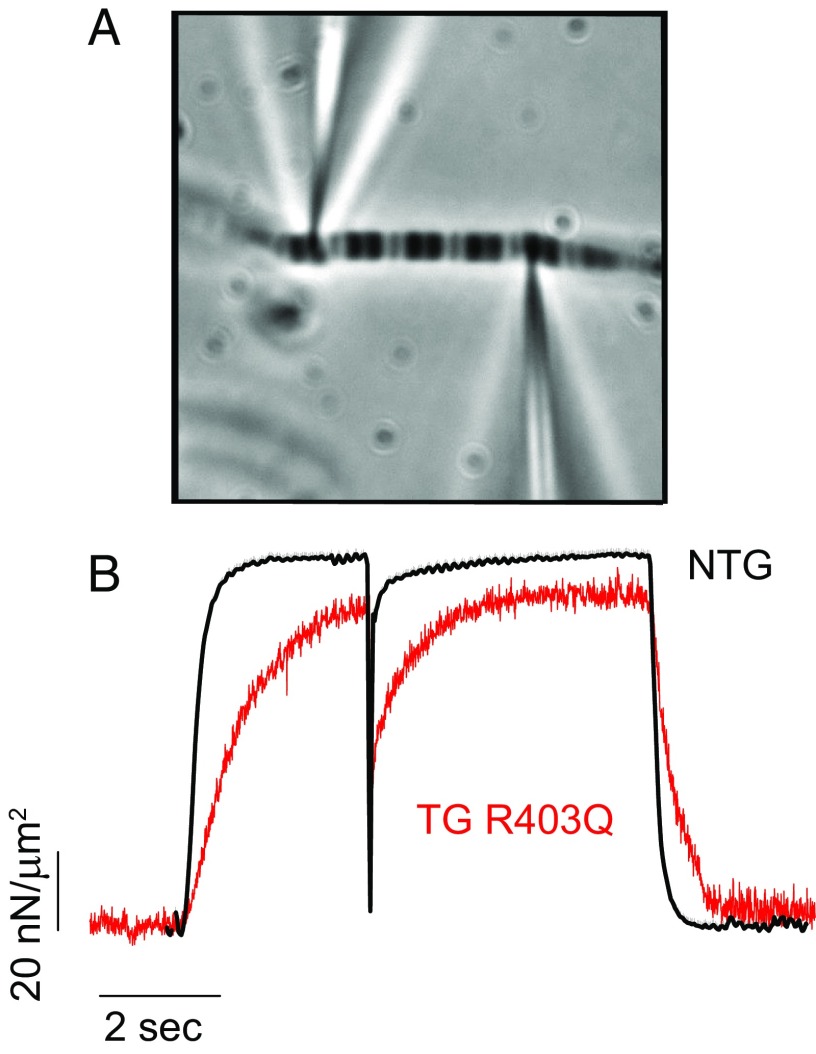
Representative traces of experiments performed with isolated myofibrils from the ventricles of TG and NTG rabbits are shown in Fig. 5. Note that due to the newly developed atomic force microscope used in these experiments (27, 28), the force traces present a clear pattern and a high signal-to-noise ratio, which allows us to compare small changes in force development, not always possible with experiments using cardiomyocytes. The force produced by the NTG myofibril was in the same range as that observed in other studies with cardiac myofibrils (28, 29) with low levels of oscillations after stabilization.
Fig. 5.
Kinetics for single rabbit myofibrils. (A) Picture of a striated myofibril attached between two microneedles in an experimental bath. (B) Representative force traces collected during experiments conducted with myofibrils isolated from NTG and TG ventricles. After myofibrils were fully activated, a shortening-stretch protocol was imposed, during which the force dropped and then redeveloped to a new steady state. The force was smaller in the TG myofibril. The rates of force development during initial activation (kact), force redevelopment after shortening-stretch (ktr), and relaxation (krel) were slower in the TG myofibril. See also SI Appendix, Fig. S1.
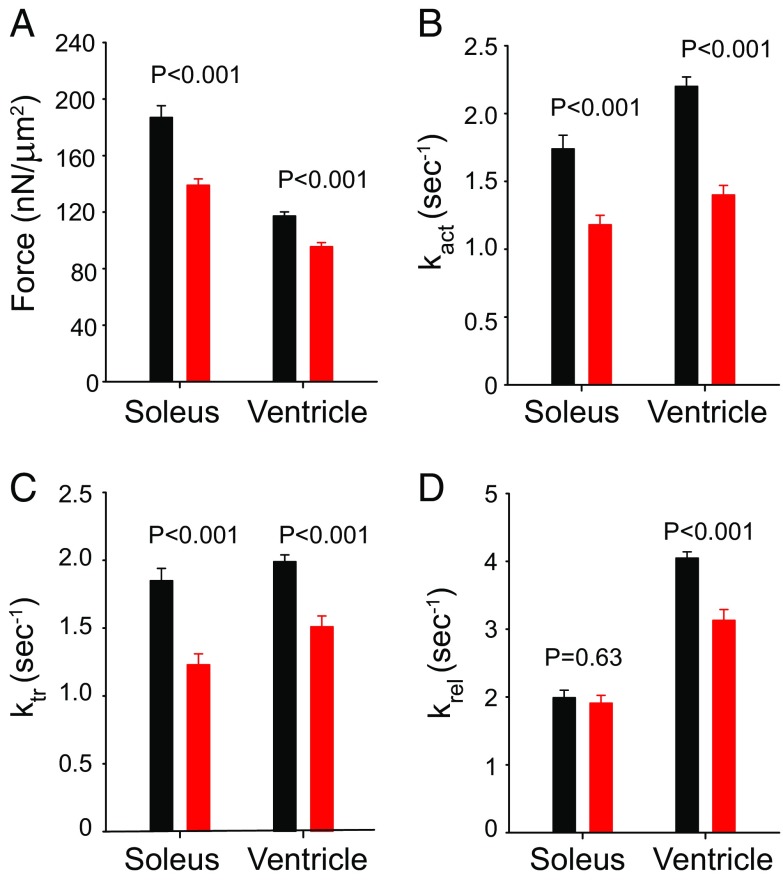
The isometric force produced by the TG (R403Q) myofibril was lower than the force produced by the NTG myofibril, before or after a shortening was imposed on the preparation. The results from one such experiment in Fig. 5 were confirmed statistically when all tested myofibrils are grouped (Fig. 6A): the force produced by the TG myofibrils was significantly lower than the NTG myofibrils. During activation and relaxation of the myofibrils, the force development and relaxation were slower in the TG myofibrils compared with the NTG myofibrils (Fig. 6), and the TG myofibril reached a steady-state plateau in force later than the NTG myofibril. As a result the rates of activation (kact) and relaxation (krel) were significantly slower in the mutants than in the control myofibrils (Fig. 6 B and D).
Fig. 6.
Mean values (±SEM) of (A) force, (B) kact, (C) ktr, and (D) krel obtained with soleus and ventricle myofibrils isolated from NTG (black) and TG (red) muscles. Statistical P values were obtained after t tests comparing the two groups of myofibrils in each condition. The sample sizes were as follows. Soleus: 8 NTG rabbits (63 experiments); 9 TG rabbits (68 experiments). Ventricles: 8 NTG rabbits (56 experiments); 9 TG rabbits (71 experiments). Details are in SI Appendix, Table S2.
After full activation was obtained, the myofibrils underwent a fast shortening protocol for measurements of force redevelopment (ktr), a putative indicator of cross-bridge kinetics (30) that has been also used with cardiac myofibrils to evaluate myosin–actin interactions (28, 29). The experiment depicted in Fig. 5 clearly shows that the force redevelopment is slower in the TG myofibril than in the NTG myofibril, a result that was consistently observed in our study with all myofibrils (Fig. 6C). These results suggest that cross-bridge kinetics are affected in the TG myofibrils, such that the redevelopment of force is slowed.
The results for soleus myofibrils are virtually the same as for the ventricle myofibrils, and most of the parameters that we measured during the experiments were decreased in the TG samples compared with the NTG samples: the isometric force was lower, and the kact and ktr were slower (Fig. 6 A–C). The only parameter that was not changed in the TG soleus myofibrils was the krel (Fig. 6D). The force produced by the soleus is higher than the force produced by the ventricles, consistent with previous studies (31, 32)
The Power Output in R403Q Mutant Myofibrils Is Significantly Reduced Compared with NTG Controls.
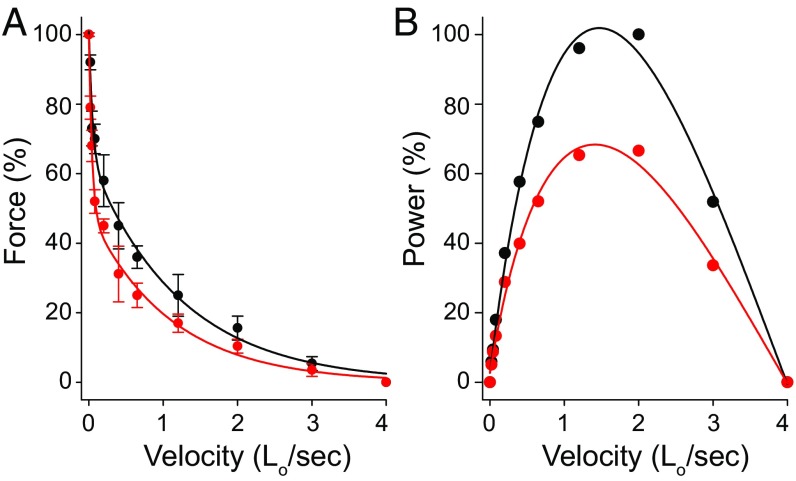
In some ventricle myofibrils (n = 20 for each group), we performed (i) slack tests to measure the maximum velocity of shortening, and (ii) contractions in which the myofibrils were released during maximal activation by predetermined step reductions in length (0.007 SL/s to 4 SL/s) to construct a force–velocity and power–velocity relation. During the slack tests the myofibrils were allowed to reach maximal force and a length step was imposed rapidly (2 ms), allowing the myofibril to become slack and the tension to drop to zero. The time required for the myofibril to redevelop tension relative to the imposed length steps was fitted with a first-order least-squares regression line, and the corresponding slope was used as a measurement of the unloaded shortening velocity. During the step reductions in length, the force transiently drops to zero before taking up the shortening. The force at the end of shortening was fitted using Hill’s equation: (F + a) × (V + b) = b (Fo + a) (see details in SI Appendix, SI Materials and Methods). The maximum velocity of shortening was decreased in the TG myofibrils. When normalized per maximum force produced by individual myofibrils, the entire force–velocity relationship obtained for the TG mutant showed a leftward shift compared with the NTG myofibrils (Fig. 7A), suggesting that less force is produced for a given velocity of contraction. Such a shift results in a diminished power output (F × V) in the TG myofibrils (Fig. 7B).
Fig. 7.
Force–velocity relation (A) obtained with myofibrils isolated from NTG (black) and TG (red) ventricles. The curves depicted with solid lines were obtained by fitting the data with the Hill equation (v + b) (F + a) = b (Fo + a), where F is force, v is velocity, Fo is the maximal isometric force, a and b are constants. There is a downward shift in the curve for the TG myofibrils. (B) Power–velocity curve obtained with myofibrils isolated from NTG and TG ventricles. The power is smaller in TG myofibrils. The values for the Hill equation are given in SI Appendix, Table S3. Curves before normalization are shown in SI Appendix, Fig. S2.
Discussion
The unique contribution reported here is the use of a large animal model, the TG rabbit, to characterize the effect of the R403Q mutation in the β-MHC on actin–myosin interactions at the molecular and myofibrillar levels. We find that the ATPase activity of R403Q myosin S1 is similar to that of the S1 control, but the in vitro actin motility is markedly reduced for myosin containing the R403Q mutation in only one of its two heavy chains. Importantly, single R403Q myofibrils, which represent the smallest contractile unit with a 3D lattice containing all of the major proteins, showed reduced rates for activation and relaxation, and a reduced force and velocity profile leading to less power output from the heart.
Hypertrophic Cardiomyopathy Is Primarily a Disease of Cardiac Myocytes.
Beginning with the discovery of the R403Q mutation, causal mutations in more than a dozen genes have been found for all of the major proteins of the cardiac sarcomere as well as in the adjacent Z-disk (reviewed in ref. 33). Mutations in the genes for MHC (MYH7) and for myosin-binding protein C (MYBPC3) account for more than half of the patients with this complex cardiovascular disease (34). The majority of the causal missense mutations are in MYH7 (>300), whereas truncation mutations are prevalent in MYBPC3 (35). Environmental factors and so-called “modifier genes” (genetic background) can also lead to cardiac hypertrophy, and these patients serve as an important control in studies on the effects of “causal genes” in human hearts. Recent studies on the mechanics of single myofibrils from patients with the R403Q mutation found a significant change in force for only one of three patients (29, 36), highlighting the difficulties in understanding the consequences for even a single severe mutation. Although studies on human tissue are obviously invaluable for purposes of diagnosis and improved treatment strategies, there is a strong need for a simpler animal model to increase our understanding of functional consequences caused by the mutation.
The creation of a TG rabbit made it possible to study the effect of a point mutation in a β-MHC backbone at the molecular and myofibrillar levels without the extensive cardiac remodeling that obscures the primary cause of HCM in humans. We find a loss-of-function in the rabbit model similar to our more limited findings in the TG mouse model (20, 21). Furthermore, we conclude that many of the contradictions in the literature regarding the function of a single missense mutation are related to myosin isoform dependence. But the small sequence variations in the cardiac β-myosin isoform across different animal species appear to have less impact on the phenotype of a causal mutation than expression in an α-myosin backbone of the same species (37).
Molecular Characterization of Mutant β-Myosin.
We have previously shown that the R403Q mutation in a mouse β-backbone produces very little change in function by steady-state and transient kinetics of S1, in contrast to the enhanced function in mouse cardiac α-myosin (20, 21). Because disease phenotypes are small and, in fact, close to experimental error in biochemical/biophysical experiments, we believed it essential to demonstrate functional aspects in a larger mammal before reaching any firm conclusions about the effect of a specific mutation. The simplest large-animal model that has been previously studied was the TG rabbit with the R403Q mutation cloned into a human heavy chain (22). Here we generated a similar rabbit, but the R403Q was cloned into the rabbit heavy chain to preserve a homologous system. In contrast to the first rabbit studies (22), in which the 2-y survival rate was reduced by ∼50%, our rabbits showed no symptoms of disease or premature death over this period. The difference may be due to the much higher R403Q protein expression level in the first rabbit line, 41% by 2D isoelectric focusing gels, which can separate human from rabbit myosin proteins. The mismatch caused by the introduction of the human myosin into a rabbit heart may also have contributed to the disease phenotype, although the 2-y survival rate for the wild-type rabbits was near normal (22).
A consistent finding between the TG mouse and the TG rabbit is that the R403Q mutation in β-MHC can cause a decrease in function, but never an increase. In the TG rabbit, the endogenous β-cardiac background made it possible to measure the in vitro motility of whole myosin, which was markedly reduced by the R403Q mutation. This raises the question of whether the earlier in vitro motility findings for human β-myosin isolated from cardiac or soleus muscle biopsies from as many as nine patients with the R403Q mutation (3, 4) are relevant to the more recent findings in animal models. The unusually large (70–80%) decrease in velocity with R403Q myosin compared with controls raised the issue of whether the loss of movement in the largely heterodimeric myosin molecules (only one heavy chain is mutated in heterozygotes) was due to ATP-insensitive “rigor-like” actomyosin linkages (38). Nonetheless, myosins from 30 hypertrophic cardiomyopathy patients containing six different mutations in the MHC, in addition to R403Q, all showed a reduction in actin velocity compared with the controls, although the majority of mutations had a smaller effect than R403Q (3). However, in a subsequent study (5) in which myosin from the same R403Q patients was extensively pretreated with unlabeled actin to remove the contribution of noncycling heads, the investigators found a small enhancement in actin filament motility (5). These contrasting results underscore the drawbacks of studies with heterogeneous patient samples.
The inability to express striated muscle myosin was overcome recently by the development of an adenoviral mediated myogenic expression system (C2C12) for human β-cardiac myosin (39, 40). Several HCM mutants have been expressed using a short human heavy chain bound to ventricular ELC (sS1). Most of the mutations, whether near the nucleotide binding site, R453C (13), the R403Q site described here (12), or in the converter region—R719W, R723G, G741R (11)—produced small or no effects on motor function, despite the expectation of a gain-of-function based on the clinical hypercontractility. The extensive study on the properties of R403Q sS1 with both pure and tropomyosin, troponin-regulated actin concluded that the data fit best with a loss of contractility under loaded conditions (12).
The recombinant human R403Q S1 characterized by Nag et al. (12) and the rabbit R403Q S1 described here have in common that each shows an ATPase activity and Km that are similar to their wild-type controls. The unloaded in vitro velocity for the expressed human S1 showed no significant difference between the mutant and wild-type sS1, but single-headed S1 with a truncated lever arm (no RLC) is expected to be less discriminating in motility experiments than full-length myosin.
Mechanical Properties of Single Myofibrils Containing a β-Myosin Mutation.
The basic organized structural unit in a muscle fiber is the myofibril. The two major proteins, actin and myosin, are assembled into filamentous structures whose interaction during contraction and relaxation is regulated by a host of proteins associated with the thin (actin) and thick (myosin) filaments. To minimize the effect of the associated proteins on the phenotype caused by the R403Q mutation, we characterized myofibrils from cardiac and soleus rabbit muscles, both expressing the identical β-myosin from the MYH7 gene, but containing muscle-specific isoforms for the other proteins. We find that the maximum tension and the kinetic rates for activation, redevelopment of tension, and relaxation are all reduced in the ventricular R403Q myofibrils relative to controls, and similarly in the soleus rabbit myofibrils with the exception of the rate for relaxation.
Data on myofibrils from human patients with sarcomeric mutations are limited, but two recent studies stand out insofar as they tried to distinguish the effect of cardiac remodeling from the intrinsic mutation in inherited cardiac hypertrophy. A study on single cardiomyocytes and myofibrils from patients with different mutations in the MHY7 gene found that their maximal force-generating capacity was significantly reduced compared with patients with impaired cardiac function, but no discernable sarcomeric mutations (41). However, some of the loss in force was attributed to cardiac remodeling, because tissue from nonfailing donor hearts had the highest force production. A separate study on three patients with the R403Q mutation, one of whom had been characterized previously in myofibril kinetics (29), found a significant increase in cross-bridge relaxation kinetics with the cardiac myofibrils, but at a greater energetic cost in tension generation as measured in muscle strips (36). These kinetic results are in contrast to our present study, but the reasons for such differences can involve many factors, including the varying clinical conditions of the patients, the different degrees of heart remodeling, and the methods used for isolating and testing myofibrils.
Our results showing a downward shift in the force–velocity relation further strengthens the conclusion of a loss-of-function in the myofibrils affected by the mutation. We observed that the R403Q myofibrils produced less power (F × V) than the control myofibrils (i.e., the loss in function is ultimately a decreased power output from the ventricles). We recognize that the transgenic rabbit is “only a model” and all models have limitations (42). But the closer resemblance of a rabbit heart to a human heart offers fundamental insights into the mechanistic consequences of a single missense mutation in β-cardiac (slow skeletal) myosin.
Conclusions and Perspectives
Based on our functional studies with the R403Q mutation in a rabbit model, we conclude that this long-studied mutation, for which the “cardiomyopathy loop” (CM-loop) was named, most likely leads to a loss-of-function at the molecular and myofibrillar levels. But this result should not necessarily be extrapolated to the countless other mutations in myosin. The R403 residue is highly conserved in all class II myosins, and is located at the base of a surface loop that forms a major hydrophobic interface with actin (Fig. 1). The recent structures obtained at near atomic resolution by electron cryomicroscopy have shown that the R403 residue interacts with neighboring residues to stabilize the conformation of the CM-loop (43). Therefore, although a mutation at this site can be expected to have a large impact on the actomyosin interaction, it may also destabilize the sequestered, folded heads of myosin in the relaxed state (44). With increasing knowledge and advanced techniques, we have come to realize that understanding the molecular basis for this complex heart disease is far more difficult than previously anticipated, and will require extensive future studies in large animal models, and expression of full-length cardiac myosin, to arrive at a unifying mechanism.
Materials and Methods
All methods, including construction of the transgenic rabbit, biochemical procedures, and myofibril preparation and mechanical measurements, are included in SI Appendix. All animal experiments were conducted in accordance with the Guide for the Care and Use of Laboratory Animals (45) and approved by the Institutional Animal Care and Use Committee at Cincinnati Children’s Hospital.
Supplementary Material
Acknowledgments
This study was supported by NIH Grants R01 AR053975 and R21 HL111847 (to S.L.), HL110869 (to K.M.T.), and 2P01 HL69779 and R01 HL056370 (to J.R.); and the Canadian Institutes for Health Research and the Natural Science and Engineering Research Council of Canada (D.E.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1802967115/-/DCSupplemental.
References
- 1.Geisterfer-Lowrance AA, et al. A molecular basis for familial hypertrophic cardiomyopathy: A beta cardiac myosin heavy chain gene missense mutation. Cell. 1990;62:999–1006. doi: 10.1016/0092-8674(90)90274-i. [DOI] [PubMed] [Google Scholar]
- 2.Lowey S. Functional consequences of mutations in the myosin heavy chain at sites implicated in familial hypertrophic cardiomyopathy. Trends Cardiovasc Med. 2002;12:348–354. doi: 10.1016/s1050-1738(02)00181-0. [DOI] [PubMed] [Google Scholar]
- 3.Cuda G, Fananapazir L, Epstein ND, Sellers JR. The in vitro motility activity of beta-cardiac myosin depends on the nature of the beta-myosin heavy chain gene mutation in hypertrophic cardiomyopathy. J Muscle Res Cell Motil. 1997;18:275–283. doi: 10.1023/a:1018613907574. [DOI] [PubMed] [Google Scholar]
- 4.Cuda G, Fananapazir L, Zhu WS, Sellers JR, Epstein ND. Skeletal muscle expression and abnormal function of beta-myosin in hypertrophic cardiomyopathy. J Clin Invest. 1993;91:2861–2865. doi: 10.1172/JCI116530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palmiter KA, et al. R403Q and L908V mutant beta-cardiac myosin from patients with familial hypertrophic cardiomyopathy exhibit enhanced mechanical performance at the single molecule level. J Muscle Res Cell Motil. 2000;21:609–620. doi: 10.1023/a:1005678905119. [DOI] [PubMed] [Google Scholar]
- 6.Yamashita H, Tyska MJ, Warshaw DM, Lowey S, Trybus KM. Functional consequences of mutations in the smooth muscle myosin heavy chain at sites implicated in familial hypertrophic cardiomyopathy. J Biol Chem. 2000;275:28045–28052. doi: 10.1074/jbc.M005485200. [DOI] [PubMed] [Google Scholar]
- 7.Fujita H, et al. Characterization of mutant myosins of Dictyostelium discoideum equivalent to human familial hypertrophic cardiomyopathy mutants. Molecular force level of mutant myosins may have a prognostic implication. J Clin Invest. 1997;99:1010–1015. doi: 10.1172/JCI119228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sata M, Ikebe M. Functional analysis of the mutations in the human cardiac beta-myosin that are responsible for familial hypertrophic cardiomyopathy. Implication for the clinical outcome. J Clin Invest. 1996;98:2866–2873. doi: 10.1172/JCI119115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sweeney HL, Straceski AJ, Leinwand LA, Tikunov BA, Faust L. Heterologous expression of a cardiomyopathic myosin that is defective in its actin interaction. J Biol Chem. 1994;269:1603–1605. [PubMed] [Google Scholar]
- 10.Adhikari AS, et al. Early-onset hypertrophic cardiomyopathy mutations significantly increase the velocity, force, and actin-activated ATPase activity of human β-cardiac myosin. Cell Rep. 2016;17:2857–2864. doi: 10.1016/j.celrep.2016.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawana M, Sarkar SS, Sutton S, Ruppel KM, Spudich JA. Biophysical properties of human β-cardiac myosin with converter mutations that cause hypertrophic cardiomyopathy. Sci Adv. 2017;3:e1601959. doi: 10.1126/sciadv.1601959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nag S, et al. Contractility parameters of human β-cardiac myosin with the hypertrophic cardiomyopathy mutation R403Q show loss of motor function. Sci Adv. 2015;1:e1500511. doi: 10.1126/sciadv.1500511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sommese RF, et al. Molecular consequences of the R453C hypertrophic cardiomyopathy mutation on human β-cardiac myosin motor function. Proc Natl Acad Sci USA. 2013;110:12607–12612. doi: 10.1073/pnas.1309493110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geisterfer-Lowrance AA, et al. A mouse model of familial hypertrophic cardiomyopathy. Science. 1996;272:731–734. doi: 10.1126/science.272.5262.731. [DOI] [PubMed] [Google Scholar]
- 15.Moore JR, Leinwand L, Warshaw DM. Understanding cardiomyopathy phenotypes based on the functional impact of mutations in the myosin motor. Circ Res. 2012;111:375–385. doi: 10.1161/CIRCRESAHA.110.223842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tyska MJ, et al. Single-molecule mechanics of R403Q cardiac myosin isolated from the mouse model of familial hypertrophic cardiomyopathy. Circ Res. 2000;86:737–744. doi: 10.1161/01.res.86.7.737. [DOI] [PubMed] [Google Scholar]
- 17.Palmer BM, et al. Myofilament mechanical performance is enhanced by R403Q myosin in mouse myocardium independent of sex. Am J Physiol Heart Circ Physiol. 2008;294:H1939–H1947. doi: 10.1152/ajpheart.00644.2007. [DOI] [PubMed] [Google Scholar]
- 18.Alpert NR, et al. Molecular mechanics of mouse cardiac myosin isoforms. Am J Physiol Heart Circ Physiol. 2002;283:H1446–H1454. doi: 10.1152/ajpheart.00274.2002. [DOI] [PubMed] [Google Scholar]
- 19.Malmqvist UP, Aronshtam A, Lowey S. Cardiac myosin isoforms from different species have unique enzymatic and mechanical properties. Biochemistry. 2004;43:15058–15065. doi: 10.1021/bi0495329. [DOI] [PubMed] [Google Scholar]
- 20.Lowey S, et al. Functional effects of the hypertrophic cardiomyopathy R403Q mutation are different in an alpha- or beta-myosin heavy chain backbone. J Biol Chem. 2008;283:20579–20589. doi: 10.1074/jbc.M800554200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowey S, Bretton V, Gulick J, Robbins J, Trybus KM. Transgenic mouse α- and β-cardiac myosins containing the R403Q mutation show isoform-dependent transient kinetic differences. J Biol Chem. 2013;288:14780–14787. doi: 10.1074/jbc.M113.450668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marian AJ, et al. A transgenic rabbit model for human hypertrophic cardiomyopathy. J Clin Invest. 1999;104:1683–1692. doi: 10.1172/JCI7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagueh SF, et al. Evolution of expression of cardiac phenotypes over a 4-year period in the beta-myosin heavy chain-Q403 transgenic rabbit model of human hypertrophic cardiomyopathy. J Mol Cell Cardiol. 2004;36:663–673. doi: 10.1016/j.yjmcc.2004.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanbe A, et al. Transgenic rabbit model for human troponin I-based hypertrophic cardiomyopathy. Circulation. 2005;111:2330–2338. doi: 10.1161/01.CIR.0000164234.24957.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chizzonite RA, et al. Isolation and characterization of two molecular variants of myosin heavy chain from rabbit ventricle. Change in their content during normal growth and after treatment with thyroid hormone. J Biol Chem. 1982;257:2056–2065. [PubMed] [Google Scholar]
- 26.Waller GS, Ouyang G, Swafford J, Vibert P, Lowey S. A minimal motor domain from chicken skeletal muscle myosin. J Biol Chem. 1995;270:15348–15352. doi: 10.1074/jbc.270.25.15348. [DOI] [PubMed] [Google Scholar]
- 27.Labuda A, Brastaviceanu T, Pavlov I, Paul W, Rassier DE. Optical detection system for probing cantilever deflections parallel to a sample surface. Rev Sci Instrum. 2011;82:013701. doi: 10.1063/1.3527913. [DOI] [PubMed] [Google Scholar]
- 28.Ribeiro PA, et al. Contractility of myofibrils from the heart and diaphragm muscles measured with atomic force cantilevers: Effects of heart-specific deletion of arginyl-tRNA-protein transferase. Int J Cardiol. 2013;168:3564–3571. doi: 10.1016/j.ijcard.2013.05.069. [DOI] [PubMed] [Google Scholar]
- 29.Belus A, et al. The familial hypertrophic cardiomyopathy-associated myosin mutation R403Q accelerates tension generation and relaxation of human cardiac myofibrils. J Physiol. 2008;586:3639–3644. doi: 10.1113/jphysiol.2008.155952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brenner B, Chalovich JM, Greene LE, Eisenberg E, Schoenberg M. Stiffness of skinned rabbit psoas fibers in MgATP and MgPPi solution. Biophys J. 1986;50:685–691. doi: 10.1016/S0006-3495(86)83509-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stehle R, Krüger M, Pfitzer G. Force kinetics and individual sarcomere dynamics in cardiac myofibrils after rapid ca(2+) changes. Biophys J. 2002;83:2152–2161. doi: 10.1016/S0006-3495(02)73975-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stehle R, Solzin J, Iorga B, Poggesi C. Insights into the kinetics of Ca2+-regulated contraction and relaxation from myofibril studies. Pflugers Arch. 2009;458:337–357. doi: 10.1007/s00424-008-0630-2. [DOI] [PubMed] [Google Scholar]
- 33.Maron BJ, Maron MS. Hypertrophic cardiomyopathy. Lancet. 2013;381:242–255. doi: 10.1016/S0140-6736(12)60397-3. [DOI] [PubMed] [Google Scholar]
- 34.Marian AJ. Genetic determinants of cardiac hypertrophy. Curr Opin Cardiol. 2008;23:199–205. doi: 10.1097/HCO.0b013e3282fc27d9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Helms AS, et al. Sarcomere mutation-specific expression patterns in human hypertrophic cardiomyopathy. Circ Cardiovasc Genet. 2014;7:434–443. doi: 10.1161/CIRCGENETICS.113.000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Witjas-Paalberends ER, et al. Faster cross-bridge detachment and increased tension cost in human hypertrophic cardiomyopathy with the R403Q MYH7 mutation. J Physiol. 2014;592:3257–3272. doi: 10.1113/jphysiol.2014.274571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pellegrino MA, et al. Orthologous myosin isoforms and scaling of shortening velocity with body size in mouse, rat, rabbit and human muscles. J Physiol. 2003;546:677–689. doi: 10.1113/jphysiol.2002.027375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Homsher E, Wang F, Sellers JR. Factors affecting movement of F-actin filaments propelled by skeletal muscle heavy meromyosin. Am J Physiol. 1992;262:C714–C723. doi: 10.1152/ajpcell.1992.262.3.C714. [DOI] [PubMed] [Google Scholar]
- 39.Deacon JC, Bloemink MJ, Rezavandi H, Geeves MA, Leinwand LA. Identification of functional differences between recombinant human α and β cardiac myosin motors. Cell Mol Life Sci. 2012;69:2261–2277. doi: 10.1007/s00018-012-0927-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Srikakulam R, Winkelmann DA. Chaperone-mediated folding and assembly of myosin in striated muscle. J Cell Sci. 2004;117:641–652. doi: 10.1242/jcs.00899. [DOI] [PubMed] [Google Scholar]
- 41.Witjas-Paalberends ER, et al. Mutations in MYH7 reduce the force generating capacity of sarcomeres in human familial hypertrophic cardiomyopathy. Cardiovasc Res. 2013;99:432–441. doi: 10.1093/cvr/cvt119. [DOI] [PubMed] [Google Scholar]
- 42.Marian AJ. Modeling human disease phenotype in model organisms: “It’s only a model!”. Circ Res. 2011;109:356–359. doi: 10.1161/CIRCRESAHA.111.249409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.von der Ecken J, Heissler SM, Pathan-Chhatbar S, Manstein DJ, Raunser S. Cryo-EM structure of a human cytoplasmic actomyosin complex at near-atomic resolution. Nature. 2016;534:724–728. doi: 10.1038/nature18295. [DOI] [PubMed] [Google Scholar]
- 44.Nag S, et al. The myosin mesa and the basis of hypercontractility caused by hypertrophic cardiomyopathy mutations. Nat Struct Mol Biol. 2017;24:525–533. doi: 10.1038/nsmb.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.National Research Council of the National Academies . Guide for the Care and Use of Laboratory Animals. 8th Ed National Academies Press; Washington, DC: 2011. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.