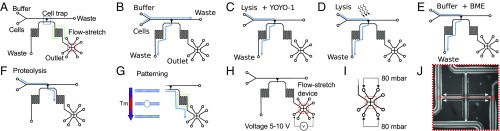
Fig. 2.
Device operation to realize the workflow shown in Fig. 1D. (A) The device comprises a main channel with inlets for cells and buffer, a cell trap connecting the main channel to the outlets. On the outlet side, a flow stretch device is placed. (B) Cells are introduced, and a single cell is captured in the trap. The flow is stopped when a cell occupies the trap. (C) Lysis buffer is introduced to remove the cytoplasm. The lysis solution contains YOYO-1 to stain the DNA of the nucleus. (D) The stained nucleus is illuminated with blue light (480 nm) to induce photonicking. (E) Buffer with BME is introduced to prevent further photonicking. (F) Same buffer to which protease K is added is introduced to release the genomic DNA by proteolysis. (G) When the DNA reaches the meandering channel, temperature is raised to partially denaturate the DNA before the device is cooled again. (H) DNA molecules are introduced to the flow-stretch device by electrophoresis and (I) stretched by a flow of buffer at a pressure of 80 mbar. (J) Brightfield image of the flow-stretch device indicating the buffer flow (dotted line) and the position of the DNA (red line). The nanoslit is 450 m across.