Fig. 4.
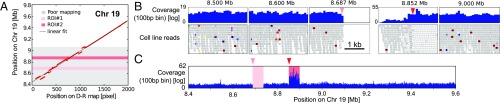
Optical D-R map and paired-end sequencing on Chr 19. (A) Plot of sliding window analysis of experimental versus in silico D-R maps, revealing a region where the experimental D-R map aligns poorly to the reference. (B) The coverage and the visual analysis of the reads in the Integrative Genomics Viewer (IGV) show three normal regions and two ROIs in this region, one is a homozygous deletion and one is a complex region. (C) The coverage of reads over the whole fragment.