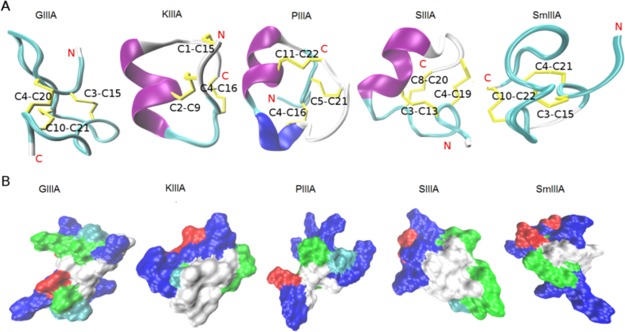
Figure 1.
Structure and surface representations of the investigated μ-conotoxins. (A) NMR structures of the five μ-conotoxins (GIIIA, KIIIA, PIIIA, SIIIA, and SmIIIA) used in this study represented as a cartoon. The secondary structure elements, α-helix (purple), 3–10 helix (blue), turn (cyan), and coil (white) were generated by STRIDE19 in visual molecular dynamics (VMD). The cysteine residues forming the disulfide bonds (yellow) were labeled. (B) Molecular surface was generated by SURF20 in VMD, indicating the hydrophobic (white), basic (blue), acidic (red), and hydrophilic (green) regions. All structures were taken from the ConoServer database.7,21