Figure 5.
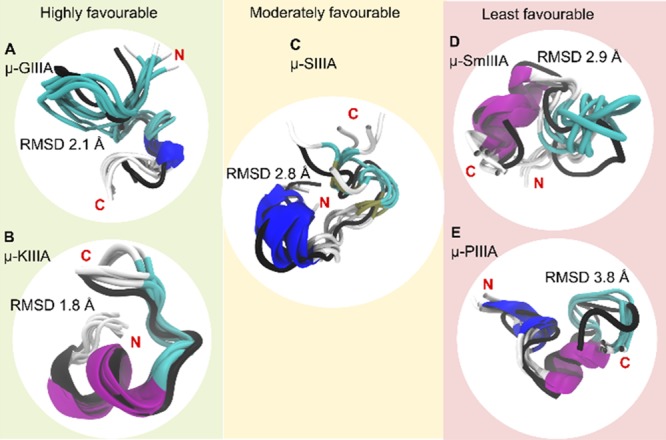
Grouping of the peptides based on the favorability of two disulfide bond stability. The C2–C5 disulfide-deficient conformations (cartoon representations colored to distinguish secondary structural elements) of the studied μ-conotoxins (A) GIIIA, (B) KIIIA, (C) SIIIA, (D) SmIIIA, and (E) PIIIA superimposed on their energy-minimized native structure (black cartoon representation). From the 300 ns trajectory, five conformations (one every 60 ns) have been used. The average RMSD of these conformations in comparison to the reference native structure is shown in Å. Besides displaying the regions of similarity and dissimilarity between the native and the C2–C5-deficient versions, the figure also provides a grouping for the five peptides in terms of favorability of the disulfide-deficient version retaining structural characteristics of the native peptide (based on RMSD, RMSF, and Rg).
