Children in a birth cohort in Dhaka, Bangladesh, were frequently infected with noroviruses that were highly genetically diverse during their first 2 years of life, both during cases of diarrhea and during monthly collection when no diarrhea was present.
Keywords: Norovirus, birth cohort study, LMIC, genotyping, evolution
Abstract
Noroviruses are a leading cause of diarrhea in children aged <5 years worldwide. We genotyped 88 viruses collected by active surveillance in a birth cohort of children <2 years of age in Dhaka, Bangladesh, during 2010–2013. Twenty-five of 31 (81%) established GI and GII genotypes were detected, with GII.4 as the predominant genotype (20%). Our results show that children in Bangladesh are infected with a great diversity of norovirus strains. Reinfections are common, but not with closely related genotypes. Birth cohort studies are critical to understand cross-protective immunity and advance the development of pediatric norovirus vaccines.
Human noroviruses are highly infectious, genetically diverse RNA viruses that are responsible for a large burden of acute gastroenteritis globally. The pathogen is known for causing severe outbreaks of foodborne disease in adults in high-income countries. Increasingly, diagnostic improvements have revealed the high burden of disease in children in low- and middle-income countries (LMICs), with an estimated 200000 deaths annually [1]. Vaccine candidates are under development, but require further understanding of the natural development of immunity [2]. Noroviruses belong to a separate genus in the family Caliciviridae, and viruses infecting humans belong to 3 genogroups (GI, GII, and GIV), which are further classified into at least 25 genotypes based on the major capsid (VP1) protein [3]. Recently, several additional GII genotypes have been proposed [4]. GII genotype 4 (GII.4) viruses are globally predominant, and new antigenic variants periodically emerge and cause global outbreaks [5]. Our understanding of the evolution of noroviruses is derived primarily from outbreak surveillance programs in high-income countries, and studies on endemic norovirus disease are limited in LMICs [6, 7], especially Asia.
In Dhaka, Bangladesh, stool samples were routinely collected from a longitudinal birth cohort of children <2 years old as part of the Etiology, Risk Factors and Interactions of Enteric Infections and Malnutrition and the Consequences for Child Health and Development (MAL-ED) study in 8 global locations [8]. Noroviruses were found to be one of the most important causes of diarrhea during the first year of life in the Bangladesh cohort, with an incidence of norovirus diarrhea of 5.5 per 100 child-months [9]. Given the high burden of norovirus observed in this population, we characterized the frequency of reinfection and the genetic diversity of noroviruses among diarrhea and nondiarrhea cases over 4 years of study (2010–2013).
METHODS
Sample Collection and Sequencing
The MAL-ED birth cohort study in Bangladesh was conducted in a slum located in the Bauniabadh area of Mirpur, Dhaka. Approval was obtained from the institutional ethics review board at the International Centre for Diarrhoeal Disease Research, Bangladesh (icddr,b). Enrollment ran from February 2010 to February 2012 using a well-defined recruitment protocol with stringent inclusion and exclusion criteria [8]. Mothers or caregivers of all participating children provided written consent at the onset, and verbal consent at each follow-up visit, with full rights to withdraw children at any time. Through community-based screening, 265 healthy newborns were enrolled within 17 days of birth and followed longitudinally until 24 months of age (Supplementary Methods). Diarrheal stools were collected within 48 hours of an episode, defined by maternal report of ≥3 loose stools in 24 hours, or 1 loose stool with visible blood. Routine nondiarrheal stools were collected monthly from age 1–12 months and quarterly at 15, 18, 21, and 24 months. All diarrheal stools and 10% of randomly selected surveillance stools were tested for norovirus (n = 2194 total stool samples tested; Supplementary Figure 1). Norovirus detection and genogroup identification (GI and GII) was performed by reverse-transcription polymerase chain reaction (RT-PCR), along with other enteropathogens using methods reported previously (Supplementary Methods). Information about norovirus incidence in the cohort by genogroup and symptoms is provided in Supplementary Table 1 and in a previous study [9].
From 447 norovirus-positive stool samples, 94 were randomly selected for genetic sequencing, based on quality and quantity of stool, norovirus cycle threshold values from RT-PCR, and confirmation that samples were free of wild-type poliovirus. The children providing these samples were representative of the total norovirus-positive population in key clinical and environmental characteristics (Supplementary Table 2). The 94 samples were sent to the US Centers for Disease Control and Prevention (CDC), where 88 were successfully sequenced, including 20 nondiarrheal and 68 diarrheal samples (Supplementary Figure 1). The CDC determined that samples were part of a nonresearch public health response, and thus institutional review board approval was not required. Partial regions encompassing the 3ʹ-end of the polymerase (RdRp) gene and the 5ʹ-end of the capsid (VP1) gene (579 bp for GI and 570 bp for GII) were amplified and sequenced using routine dual typing methods described previously [3]. GenBank accession numbers of the near-complete genome sequences of strains BG1011313 (GenBank strain name: Dhaka1928) and BG1003021 (GenBank strain name: Dhaka1882) are MG495083 and MH130046, respectively.
Data Analysis
Sequence alignments were constructed separately for GI and GII sequences, and for polymerase and capsid regions. Two reference sequences per genotype were included for comparison. Phylogenetic relationships were inferred for each of the 4 data sets using the maximum likelihood methods available in RAxML version 7.2.6 software (Supplementary Methods), incorporating a general time-reversible model of nucleotide substitution with a gamma-distributed (Γ) rate variation among sites, using the National Institutes of Health High-Performance Computing Biowulf cluster (http://hpc.nih.gov). To assess the robustness of each node, bootstrap resampling was performed (500 replicates). Immunotypes A–L were defined similarly to those proposed by Parra et al [10], as sets of phylogenetically clustered genotypes likely to be antigenically similar.
RESULTS
Frequent Detection of Norovirus in a Pediatric Cohort in Dhaka, Bangladesh
In total, 2194 of 5547 stool samples collected during 2010–2013 from the Dhaka cohort were tested for norovirus, including 1641 diarrheal and 553 nondiarrheal stool samples from 248 children aged <2 years. Of these, 325 diarrheal (19.6%) and 122 nondiarrheal (22.1%) stool samples tested positive for norovirus, representing 179 children (almost three-quarters of the cohort). Among all 447 norovirus-positive stools, 30.0% were GI, 63.3% were GII, and 6.7% were mixed GI/GII infections. Similar proportions of GI and GII were observed among monthly nondiarrheal and diarrheal stool samples (Supplementary Figure 1). A larger proportion of diarrheal samples were from older children between 12 and 24 months of age (45.2%), compared to nondiarrheal samples (28.7%). Nearly half of norovirus-positive children (n = 179) were male, 24.0% had low birth weight (<2.5 kg), and 8.4% came from households with severe food insecurity (Supplementary Table 2). The majority of children with diarrheal stools tested had at least 1 norovirus-positive stool by 12 months (51%) and 70% by 24 months. The median age at the first diarrhea-associated norovirus detection was 183 days (150 days in children who also had monthly stools tested). Children at 7 months of age were the most likely to have a norovirus-positive sample detected (Supplementary Figure 2).
Genetic Diversity of Noroviruses in a Pediatric Cohort
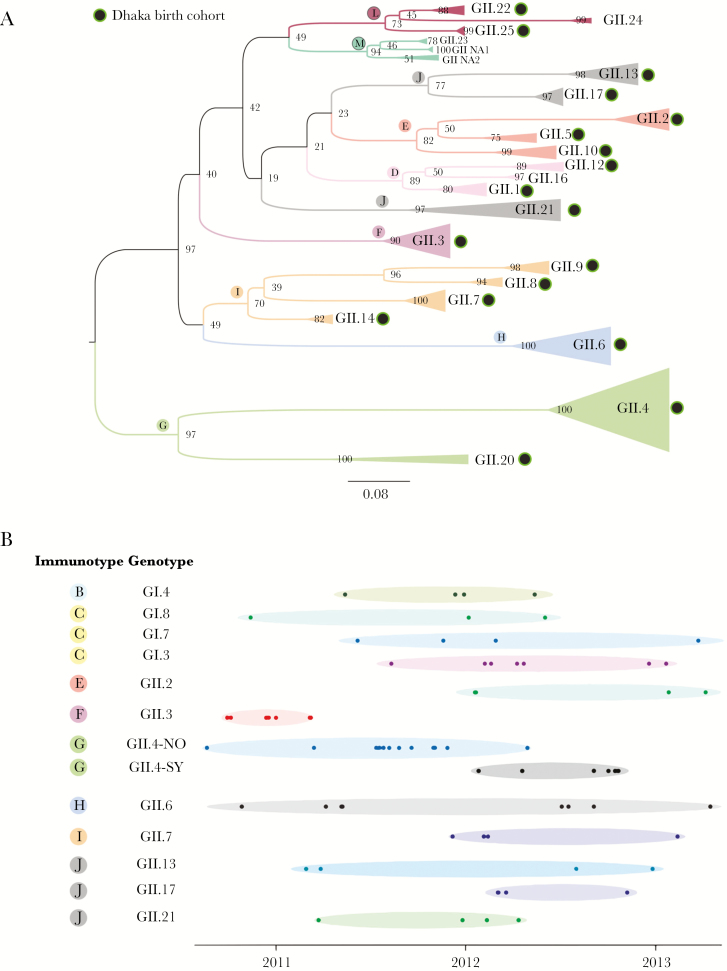
The 88 successfully sequenced samples revealed 24 different genotypes based on partial VP1 gene (see Figure 1A for GII genotypes; Supplementary Figure 3; Supplementary Table 3). No clustering of nondiarrheal vs diarrheal cases was observed on the phylogeny (Supplementary Figure 4). Six GI genotypes (GI.3–GI.8) and 18 GII genotypes (GII.1–GII.10, GII.12–GII.14, GII.17, GII.20–GII.22, and GII.25) were identified. The globally dominant GII.4 genotype was identified most frequently (~20% of sequences), including the GII.4-New Orleans (13%) and GII.4-Sydney (7%) variants. However, approximately 80% of viruses belonged to 1 of 23 other genotypes, which span almost the entire known genetic diversity of GI and GII noroviruses in humans (Figure 1A). Notably, a recently identified genotype (GII.25), which is closely related to GII.22 and GII.24, was identified in the cohort (BG1011313) [4] (Figure 1A and Supplementary Figure 3).
Figure 1.
Genetic diversity of noroviruses within a pediatric cohort in Dhaka, Bangladesh. A, Evolutionary relationships between GII noroviruses (252 nucleotide region of the 5ʹ-end of the VP1 gene) collected from children in Dhaka, with reference sequences representing genotypes observed globally included as background, inferred using maximum likelihood methods. Clades of viruses with the same genotype are represented by triangles, which are shaded according to the immunotypes A–L defined by Parra et al [10]. Bootstrap values are provided for nodes, and branch lengths are drawn to scale. The 18 GII genotypes identified in the Dhaka birth cohort are indicated with black circles with green outline. B, Each solid circle represents a norovirus sample from a child in this study. The date of collection is represented on the x-axis. Samples are ordered on the y-axis by genotype and immunotype.
In the RdRp gene, 20 polymerase genotypes (P-types) were detected (Supplementary Table 3 and Supplementary Figures 5 and 6), including 9 GI P-types (GI.P3, GI.P4, GI.P6, GI.P7, GI.P8, GI.Pa, GI.Pc, GI.Pd, and GI.Pg) and 11 GII P-types (GII.P2, GII.P4, GII.P7, GII.P16, GII.P17, GII.P20, GII.P21, GII.P22, GII.Pe, GII.Pg, and GII.Pm). The GII.P4 genotype was again predominant (18.2%). Recombination between the VP1 and RdRp gene regions was observed for both GI (n = 6 recombinant genotypes) and GII viruses (n = 12 recombinant genotypes) (Supplementary Table 4). Frequently identified recombinant genotypes included GII.P7/GII.6, GII.P16/GII.3, and GI.Pa/GI.3. No recombination events were observed between GI and GII viruses. Four GII.P17/GII.17 viruses identified in 2012 cluster with a newer lineage of GII.17 viruses that emerged in 2013 in multiple Asian countries.
Co-circulation of Genotypes Over Time
Individual genotypes were identified in the cohort at multiple time points, including those for which few (<5) viruses were available (Figure 1B). GII.6 was notable for being detected during each year of the study (2010–2013). Interactions were observed between GII.4 viruses, with GII.4-Sydney viruses replacing GII.4-New Orleans viruses in early 2012. The first detection of GII.4-Sydney in the cohort occurred on 26 January 2012 (Supplementary Table 5), in a routinely collected nondiarrheal stool, approximately 1 month before GII.4-Sydney viruses were reported in Australia in March 2012 [11]. Interactions between other genotypes were less clear, and likely require a larger sample size.
Norovirus Reinfections in the Same Child
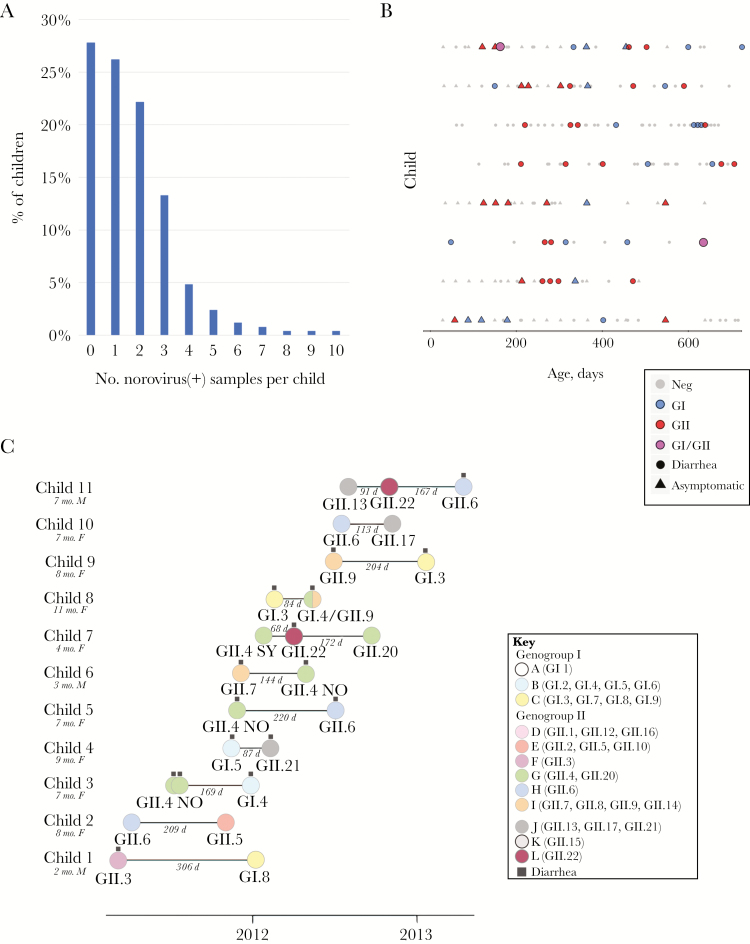
Almost half of the children in the cohort (46.0%) had >1 stool sample test positive for norovirus by PCR during the first 2 years of life (Figure 2A and Supplementary Figure 7). Of these children, the vast majority (77.2%) had 2–3 positive samples (Figure 2A). However, 1 child had as many as 10 samples test positive for norovirus during the first 2 years of life (excluding viruses samples <5 days apart that are likely to represent the same infection), and 8 children had ≥6 norovirus-positive samples (Figure 2A and 2B). Noroviruses can be shed for many weeks, and using a more stringent cutoff of 60 days between positive samples, only 2 children had >5 positive samples. Using genetic sequence data from 25 noroviruses obtained from 11 children experiencing reinfections (Figure 2C), we determined that the 11 children were reinfected with genetically distinct viruses, rather than long-term shedding of a single infection, except in the case of 2 GII.4 New Orleans viruses collected a few days apart in child 3 (Figure 2C). Reinfections typically were captured within the same year (average of 22.3 weeks between infections [range, 9.7–43.7]). No sequential infections were observed with the same genotype or “immunotype” [10]. Child 7 experienced infections with 2 immunotype G genotypes (GII.4-Sydney and GII.20), but these were separated by 240 days and not directly sequential, as a GII.22 (immunotype L) infection occurred in between.
Figure 2.
Norovirus reinfections. A, The proportion of children in the Bangladesh pediatric cohort who had a certain number (0–10) of polymerase chain reaction (PCR)–positive norovirus samples, collected at least 5 days apart, during their first 2 years of life. B, Each point represents an individual sample that was tested by PCR for norovirus, provided for the 8 children in the cohort with ≥6 PCR-positive norovirus samples, collected at least 5 days apart. The color of the point indicates the PCR result: gray, norovirus negative; blue, GI; red, GII; magenta, positive for both GI and GII. The shape of the point indicates the type of sample collection: circle, diarrheal stool; triangle, monthly nondiarrheal stool. Each row represents samples collected from an individual child, ordered from the child with the highest number of PCR-positive samples (top, n = 10) to the children with 6 PCR-positive samples (bottom). The x-axis indicates the age of the child, in days. C, Each solid circle represents a norovirus sample from a child with at least 2 samples that were genotyped for this study (n = 11 children). The age and sex of each child is provided. The color of the circle corresponds to one of the immunotypes A–L defined by Parra et al [10], similar to Figure 1. The genotype is listed below the circle. The date of sample collection is represented on the x-axis, with the number of days between sequential norovirus detections from the same child is written in gray. Circles with black squares above identify samples associated with diarrhea. Circles without black squares represent asymptomatic stools collected monthly.
DISCUSSION
The advancement of clinical trials for norovirus vaccine candidates hastens the need for greater understanding of the strength and duration of natural immunity against the diversity of norovirus strains circulating globally. In response, there have been studies of how natural herd immunity drives the cycling of norovirus genotypes within communities [12], probabilities of reinfection following GI and GII infections [9], and case studies of individuals reinfected with noroviruses of different genotypes [13]. The high viral diversity and frequent reinfection observed in Bangladeshi children suggest that larger sequencing efforts in this population could be particularly useful in understanding the epidemiology and genetic diversity of endemic, community-acquired norovirus, including the duration and strength of natural immune responses to norovirus reinfections spanning a range of antigenic distances. Even with a relatively small sampling (88 viruses), nearly the entire known diversity of noroviruses capsid genes was identified within the Bangladesh cohort, and including a newly identified genotype (GII.25). Our study aimed to characterize viral population diversity and dynamics within the cohort with minimal bias and did not specifically target reinfections from the same individual for sequencing, but our findings strongly suggest that a more targeted approach in future sequencing efforts is warranted to address natural immunity more specifically and with larger sample sizes.
At a global scale, it is possible that Dhaka and other settings with high endemic norovirus circulation serve as key viral reservoirs for genetically diverse viruses, facilitating the emergence of new antigenic variants or recombinants that become pandemic strains [14]. Retrospective surveillance in additional countries and environmental sources, including sewage [15], uncovered strains related to GII.4-Sydney and GII.17-Kawasaki viruses from years that preceded their first reports, highlighting the lingering surveillance gap between initial emergence and global detection of pandemic strains. Furthermore, while our patterns of reinfection are consistent with previously proposed immunotypes [10], larger sample sizes are required to estimate the strength and duration of immunity between diverse norovirus strains. It remains unclear whether the observed reinfections with viruses from the same immunotype (GII.4-Sydney and GII.22) suggests that the definition of immunotypes requires further refinement, or whether the intervening infection with GII.22 affected the timing of antibody waning. The high rate of asymptomatic infection of norovirus in the Dhaka birth cohort was particularly notable, and further study is needed to understand how asymptomatic infections contribute to cross-protective immunity and whether certain strains are more frequently associated with diarrhea. Sampling additional age groups also is needed to inform whether the diverse strains observed in children aged <2 years reflect other age groups, and further understanding of the age patterns of transmission could inform future vaccination strategies.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank Dr Gabriel Parra (Division of Viral Products, US Food and Drug Administration), Dr Kim Green (National Institute of Allergy and Infectious Diseases, National Institutes of Health [NIH]), and Dr Cecile Viboud (Fogarty International Center, NIH) for helpful comments on an earlier draft of this manuscript. We thank Garret Kern for his help creating figures.
Disclaimer. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the NIH, the CDC, or the US Department of Health and Human Services.
Financial support. The MAL-ED study is carried out as a collaborative project supported by the Bill & Melinda Gates Foundation, the Foundation for the NIH, and the Fogarty International Center, NIH. The icddr,b gratefully acknowledges the unrestricted support of the following donors: the government of the People’s Republic of Bangladesh, Global Affairs Canada, the Swedish International Development Cooperation Agency, and the UK Department for International Development.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Pires SM, Fischer-Walker CL, Lanata CF, et al. Aetiology-specific estimates of the global and regional incidence and mortality of diarrhoeal diseases commonly transmitted through food. PLoS One 2015; 10:e0142927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lucero Y, Vidal R, O’Ryan GM. Norovirus vaccines under development. Vaccine 2017. http://www.ncbi.nlm.nih.gov/pubmed/28668568. Accessed 10 August 2017. [DOI] [PubMed]
- 3. Vinjé J. Advances in laboratory methods for detection and typing of norovirus. J Clin Microbiol 2015; 53:373–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chhabra P, Aswath K, Collins N, et al. Near-complete genome sequences of several new norovirus genogroup II genotypes. Genome Announc 2018; 6:e00007–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lindesmith LC, Beltramello M, Donaldson EF, et al. Immunogenetic mechanisms driving norovirus GII.4 antigenic variation. PLoS Pathog 2012; 8:e1002705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lopman BA, Trivedi T, Vicuña Y, et al. Norovirus infection and disease in an Ecuadorian birth cohort: association of certain norovirus genotypes with host FUT2 secretor status. J Infect Dis 2015; 211:1813–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Saito M, Goel-Apaza S, Espetia S, et al. . Multiple norovirus infections in a birth cohort in a Peruvian periurban community. Clin Infect Dis 2014; 58:483–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Platts-Mills JA, Babji S, Bodhidatta L, et al. . Pathogen-specific burdens of community diarrhoea in developing countries: a multisite birth cohort study (MAL-ED). Lancet Glob Health 2015; 3:e564–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rouhani S, Peñataro Yori P, Paredes Olortegui M, et al. Norovirus infection and acquired immunity in 8 countries: results from the MAL-ED study. Clin Infect Dis 2016; 62:1210–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Parra GI, Squires RB, Karangwa CK, et al. Static and evolving norovirus genotypes: implications for epidemiology and immunity. PLoS Pathog 2017; 13:e1006136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Centers for Disease Control and Prevention. Emergence of new norovirus strain GII.4 Sydney—United States, 2012. MMWR Morb Mortal Wkly Rep 2013; 62:55. [PMC free article] [PubMed] [Google Scholar]
- 12. Sakon N, Yamazaki K, Nakata K, et al. Impact of genotype-specific herd immunity on the circulatory dynamism of norovirus: a 10-year longitudinal study of viral acute gastroenteritis. J Infect Dis 2015; 211:879–88. [DOI] [PubMed] [Google Scholar]
- 13. Parra GI, Green KY. Sequential gastroenteritis episodes caused by 2 norovirus genotypes. Emerg Infect Dis 2014; 20:1016–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Allen DJ, Trainor E, Callaghan A, O’Brien SJ, Cunliffe NA, Iturriza-Gómara M. Early detection of epidemic GII-4 norovirus strains in UK and Malawi: role of surveillance of sporadic acute gastroenteritis in anticipating global epidemics. PLoS One 2016; 11:e0146972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kazama S, Miura T, Masago Y, et al. Environmental surveillance of norovirus genogroups I and II for sensitive detection of epidemic variants. Appl Environ Microbiol 2017; 83:e03406–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.