Figure 2.
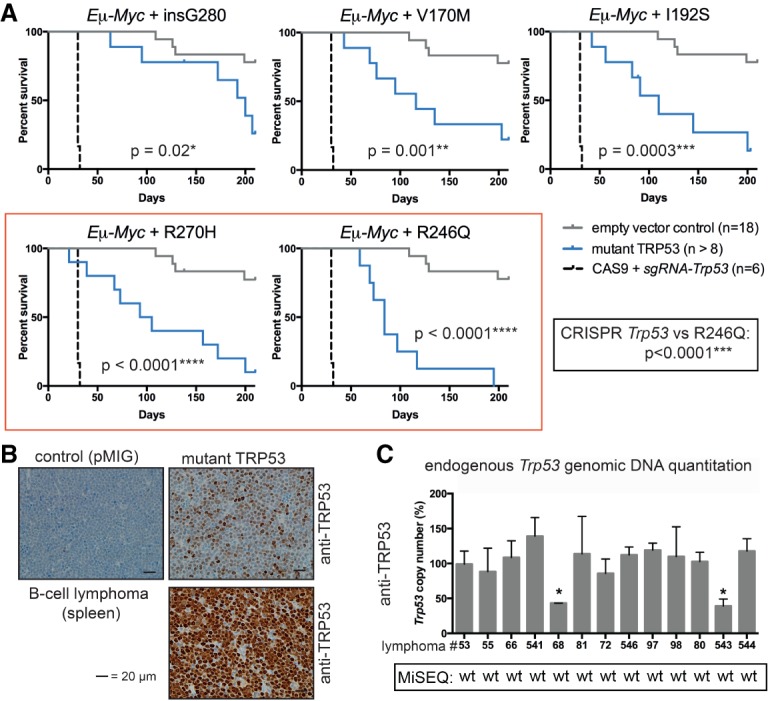
Overexpression of mutant TRP53 proteins accelerates lymphoma development in an Eμ-Myc;Trp53+/+ background and relieves selective pressure for mutation of endogenous Trp53 genes. (A) Kaplan-Meier survival curves for mice reconstituted with Eμ-Myc;Trp53+/+ HSPCs comparing empty vector control (pMIG), CRISPR/Cas9 Trp53 knockout, and each mutant TRP53 protein (V170M, I192S, G280, R246Q, and R270H). P-values were determined by log rank (Mantel-Cox) test. (B) Selected TRP53 protein immunohistochemistry in lymphomas from the Eμ-Myc hematopoietic reconstitution experiments (mice #88 and #541 plus control mouse #53). (C) Endogenous Trp53 allele copy number in lymphomas from the Eμ-Myc hematopoietic reconstitution experiments as determined by genomic DNA quantitative PCR (pMIG/control: #53; V170M: #55, #66, and #541; G280: #68 and #81; I192S: #72 and #546; R246Q: #97 and #98; R270H: #80, #543, and #544). Primary cells from Trp53−/− and Trp53+/− mice were used as controls. Data from MiSeq analysis throughout the coding region of the DNA-binding domain (exons 4–10) are indicated. (wt) Wild-type sequence. Data represent mean ± SEM. (*) P < 0.05, comparing lymphoma sample with wild-type control as determined by paired t-test.
