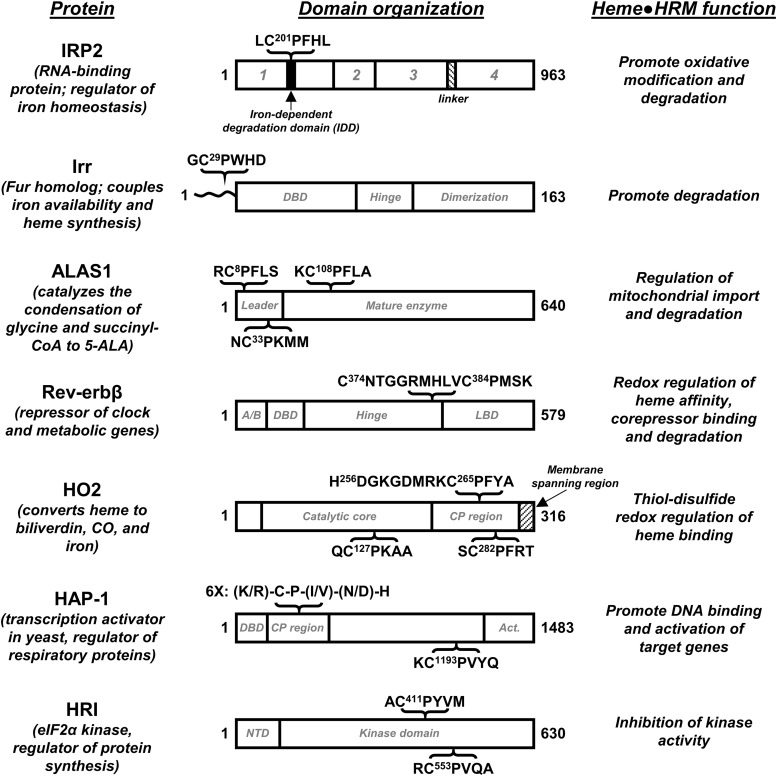
FIG. 1.
HRMs regulate the activity of enzymes, transcription factors, and an RNA regulatory protein. The modular structure of HRM-containing proteins is shown, along with the context and sequence of the HRMs. Domain organization (1–4) of IRP2 is based on its homology to aconitase (41, 74). Similarly, Irr is homologous to the Ferric Uptake Regulator (Fur) from Pseudomonas aeruginosa, and its domains are tentatively assigned as such; the N-terminal region of Irr that is distinct from Fur and contains the HRM is shown as a wavy line (29). The sequence of Rev-erbβ containing the HRM and redox-active Cys374 is shown; the A/B domain is hypervariable among members of the NR superfamily. For HO2, the sequence encompassing the heme ligand, His256 is shown in context to the nearby HRM. Hap-1 domain organization is based on functional studies (67); Act. refers to the domain that imparts transcription activation activity. NTD, or N-terminal domain of HRI is important for regulating quaternary structure (62). Delineation of domain boundaries are approximations. ALAS1, aminolevulinic acid synthase-1 nonspecific; Hap-1, heme activator protein-1; HO2, heme oxygenase isoform 2; HRM, heme regulatory motif; IRP2, iron regulatory protein 2; Irr, bacterial iron response regulator.