FIG. 2.
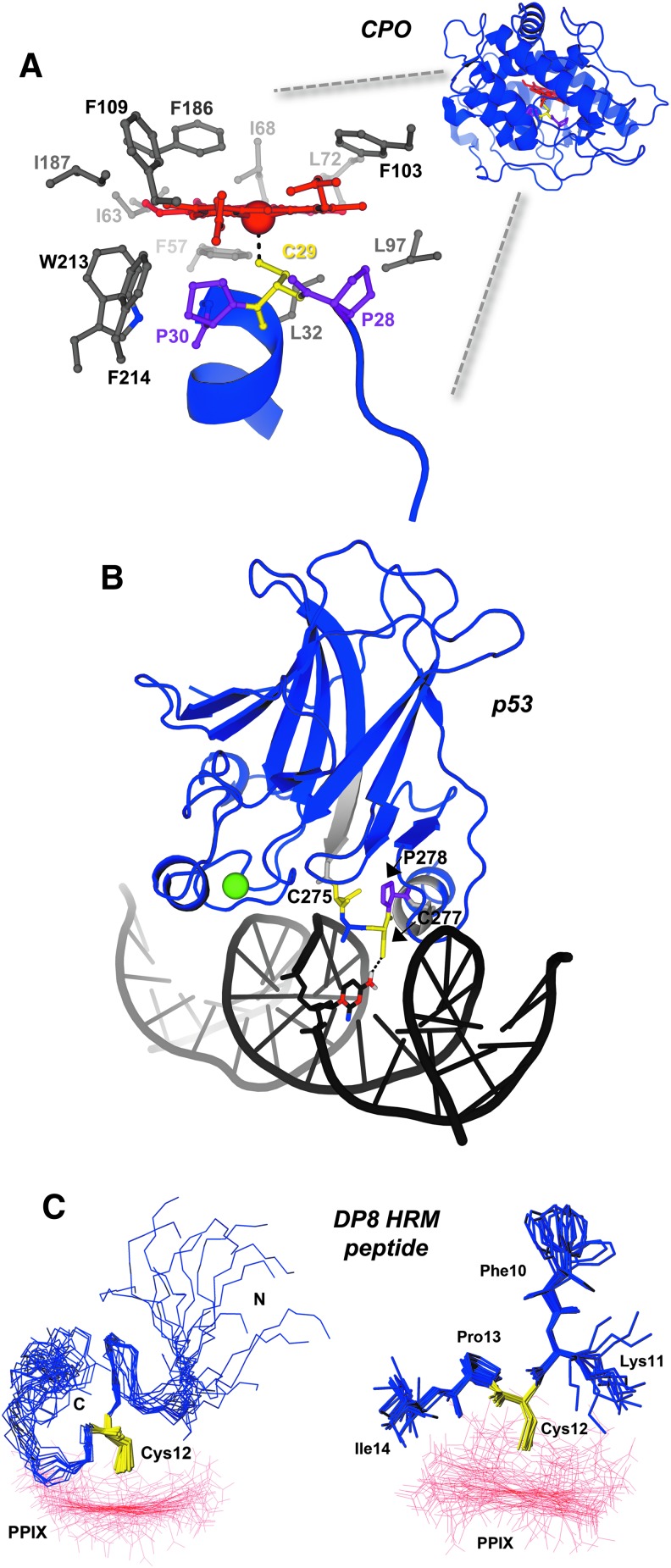
HRMs provide unique structural determinants to proteins and enzymes. (A) The overall structure of chloroperoxidase (PDB: 2ciw) shows that the HRM and Fe3+-heme cofactor are buried within the enzyme. A close-up view of the active site highlights the unique Pro-Cys-Pro HRM of CPO (prolines are shown as purple sticks, cysteine as yellow sticks), which positions the axial thiolate for heme ligation. Nonpolar residues within 4 Å of heme are shown as gray sticks. (B) p53, a transcription factor and tumor suppressor, is shown to be complexed with its cognate DNA promoter element (PDB: 1tsr). Residues flanking the HRM are shown as light gray cartoons to highlight the region, and a structural zinc atom is shown as a green sphere. (C) The NMR structure of an HRM-containing peptide from DP8 in complex with Ga3+-PPIX. Left panel, the 15 structures of lowest target function are shown with the peptide backbone as blue sticks, the HRM Cys12 in yellow sticks, and Ga3+-PPIX as red lines; the right panel depicts the inflexibility of the Cys-Pro core versus the side chains of surrounding residues. The structure serves as an excellent model demonstrating how the HRM Pro residue positions C-terminal residues away from heme, while poising the Cys-thiolate to act as a heme axial ligand. The structure of an IL-36α-derived HRM peptide in complex with Ga3+-PPIX recapitulates many of the features observed with the DP8 peptide (6). Figures in (C) are reprinted (adapted) with permission from Kühl et al. (50). Copyright (2013) American Chemical Society. CPO, chloroperoxidase; DP8, dipeptidyl peptidase 8; IL-36α, interleukin-36α; NMR, nuclear magnetic resonance; PPIX, protoporphyrin IX.