FIG. 3.
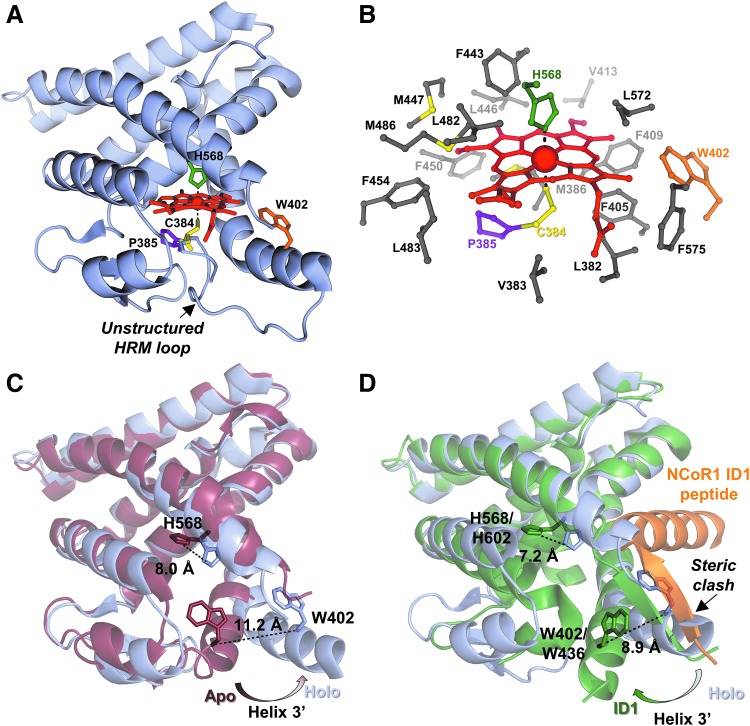
Structural insight into HRM-mediated redox regulation of heme binding to nuclear receptor, Rev-erbβ. (A) Structure of the Rev-erbβ LBD in complex with Fe3+-heme (holo-LBD, PDB: 3cqv). Heme binds in a 6-coordinate, low-spin complex with His (H568, green sticks) and Cys (C384, yellow sticks) axial ligands. C384 and P385 (purple sticks) comprise an HRM core that resides on a flexible, unstructured loop. W402 (orange sticks) provides hydrophobic contacts to heme, and its fluorescence is quenched on heme binding (unpublished observations). (B) The Rev-erbβ heme pocket is highly hydrophobic with nonpolar residues within 4 Å of heme shown as gray sticks (66). (C) Heme binding induces conformational changes of the LBD. Overlay of apo-LBD (dark red; PDB: 2v0v) and holo-LBD (light blue) structures aligned in Pymol; heme and the flexible HRM-harboring loop from the holo-LBD structure are omitted for clarity. The H3' helix swings out to accommodate the porphyrin ring, leading to a 11.2 Å shift of the W402 α-carbon. Further, conformational changes in helix 11 cause the τ-nitrogen of H568 to shift 8.0 Å, and assume its role as a Fe3+-heme ligand. (D) Pymol structural alignment of holo-LBD (light blue) versus apo-Rev-erbα LBD (dark green) in complex with an NCoR1 ID1 peptide (orange), PDB: 3n00; again, heme and the HRM loop are omitted for clarity. An anti-parallel beta sheet formed between an unraveled portion of Rev-erbα helix 11 and the NCoR1 peptide exhibits steric clash with helix H3' in the holo-LBD structure. Further, the heme axial ligand, H602 in Rev-erbα pivots away from the heme-binding pocket, suggesting that heme and NCoR1 would compete for different Rev-erb conformers. ID, interaction domain; LBD, ligand-binding domain; NCoR1, nuclear receptor compressor 1.