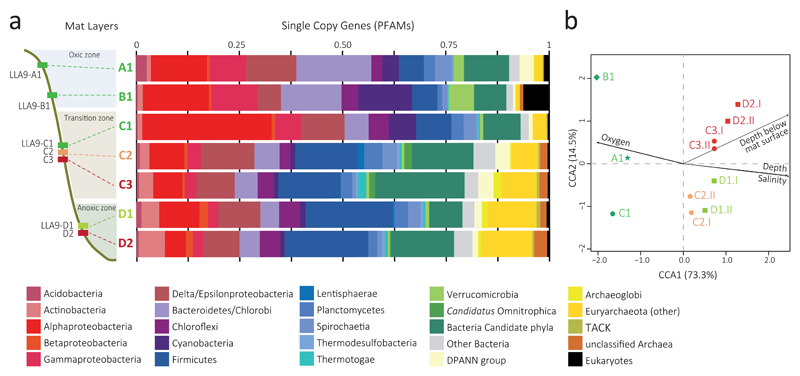
Fig. 1.
Microbial diversity in microbial mat metagenomes along a redox gradient in a pond from the Salar de Llamara (Atacama Desert, Chile). (a) Overall community composition of metagenomic assemblies inferred from the phylogenetic assignment of PFAMs corresponding to 237 single copy genes. Layers LLA9-C2, C3, D1 and D2 correspond to merged metagenomes (see Supplementary Fig. 4a for individual replicates). Relatively abundant Candidatus Omnitrophica and Archaeoglobi are highlighted independently from their respective bacterial and archaeal clades. Detailed classification of archaea and bacterial candidate divisions is shown in Supplementary Fig. 4b-c. (b) Canonical correspondence analysis (CCA) plot based on normalized COG frequencies in individual metagenome datasets showing the similarity between replicate metagenomes and how the different metagenomes correlate with local environmental conditions (see also Supplementary Fig. 5).