Abstract
Specialized queens and life-time unmated workers evolved once in the common ancestor of all ants, but whether caste development across ants continues to be at least partly regulated by a single core set of genes remains obscure. We analysed brain transcriptomes from five ant species (three subfamilies) and reconstructed the origins of genes with caste-biased expression. Ancient genes predating the Neoptera were more likely to regulate gyne (virgin queen) phenotypes, while caste differentiation roles of younger, ant-lineage-specific genes varied. Transcriptome profiling showed that the ancestral network for caste-specific gene-regulation has been maintained, but that signatures of common ancestry are obscured by later modifications. Adjusting for such differences, we identified a core gene-set that: 1. consistently displayed similar directions and degrees of caste-differentiated expression, and 2. have mostly not been reported as being involved in caste differentiation. These core regulatory genes exist in the genomes of ant species that secondarily lost the queen caste, but expression differences for reproductive and sterile workers are minor and similar to social paper wasps that lack differentiated castes. Many caste-biased ant genes have caste-differentiated expression in honeybees, but directions of caste bias were uncorrelated, as expected when permanent castes evolved independently in both lineages.
Introduction
In all but a few evolutionarily derived ants female eggs develop into either gynes (future reproductive queens) or life-time unmated workers. Such differentiated castes are comparable to germ-lines and soma, an analogy that motivated Wheeler 1 to characterize ant colonies as superorganisms. Irreversible transitions to superorganismality have also evolved in corbiculate bees, vespine wasps and evolutionary derived termites, always from ancestors where colonies have a single strictly monogamous queen, similar to obligatory multicellular eukaryotes having evolved from ancestors initiating new individuals from a single zygote 2 (reviewed by 3). Because all Metazoans have a common ancestor, their fundamental cell types are homologous, just like queen and worker castes should be homologous when all ants have a single common ancestor 4. This topic has been exhaustively explored in Metazoans 5 where homologous character development has now been confirmed to be encoded and largely regulated by the ancestral gene regulatory network (GRN), consisting of a set of orthologs that interact in shaping homologous phenotypic traits 6. However, the ontology of traits that characterize social insect superorganisms has received much less attention and evidence for the existence of GRNs that shaped their independent origins has remained ambiguous 7–9.
Detecting ancestral GRNs for superorganismal caste phenotypes is likely to be difficult, because these genes will have become extensively rewired over evolutionary time. They will also have diversified when subfamilies and genera evolved, analogous to later adjustments in metazoan developmental integration and cell type diversification 10,11. Other genes that presently affect caste differentiation and social behavior may be specific for subfamilies or genera and thus without sister lineage orthologs 12–16. Such lineage-specific genes may have facilitated new adaptations for work coordination, communication, or foraging 9,15,16, once more analogous to how we understand evolutionary elaborations across lineages of multicellular animals 17,18.
Previous studies in honeybees and ants have shown that both ancestral and novel genes have shaped caste differentiation. Many genes up-regulated in honeybee worker larvae have no Drosophila homologs, suggesting they are novel 12 similar to whole body transcriptome comparisons of adult Temnothorax longispinosus ants and larvae of Monomorium pharaonis 14,19. However, these single-species studies could not capture the diversity in life-history and social structure across species 20, and the use of whole body transcriptomes precludes detecting caste-differences at the homologous organ-level. The power of these studies to reconstruct GRNs underlying social traits therefore remained limited 21,22 compared to what has been possible for mammalian organs 23,24. The only comparative ant study so far used a whole body approach and detected only a single gene with caste-biased expression across 16 species 8. The use of whole-body transcriptomes without reference genomes 8 also implies that gene orthology across species cannot be established (Supplementary Information) and that caste-specific gene expression may seem insignificant just because differences in opposite direction across organs are averaged out 25.
The objective of this study was to assess the origin of genes with caste-biased expression and to identify the ancestral GRN in the brains of five ant species from different genera and three different subfamilies. We used re-annotated genomes to identify conserved genes and developed a method to normalize for subfamily/genus-specific effects and colony-identity before evaluating caste-biased gene-expression across ant species. This allowed us to identify a number of conserved pathways and candidate genes that have previously remained hidden. We validated our approach by examining gene expression in the brains of reproductive and sterile workers of two ants that secondarily lost the queen caste. This allowed testing the expectation that their GRNs for reproductive division of labor should be less differentiated because fertile and sterile workers represent variation within a single caste phenotype, similar to fertile and sterile females of Polistes wasps that never evolved differentiated castes with 100% predictable mating status. We also examined brain gene expression data of honeybee queens and workers to assess whether partly the same genes were involved in these two independent origins of superorganismality, and whether directionality of caste-biased expression for shared genes was correlated across the ants and bees.
Results
Reconstructing the evolutionary origin of ant genes with caste-biased expression
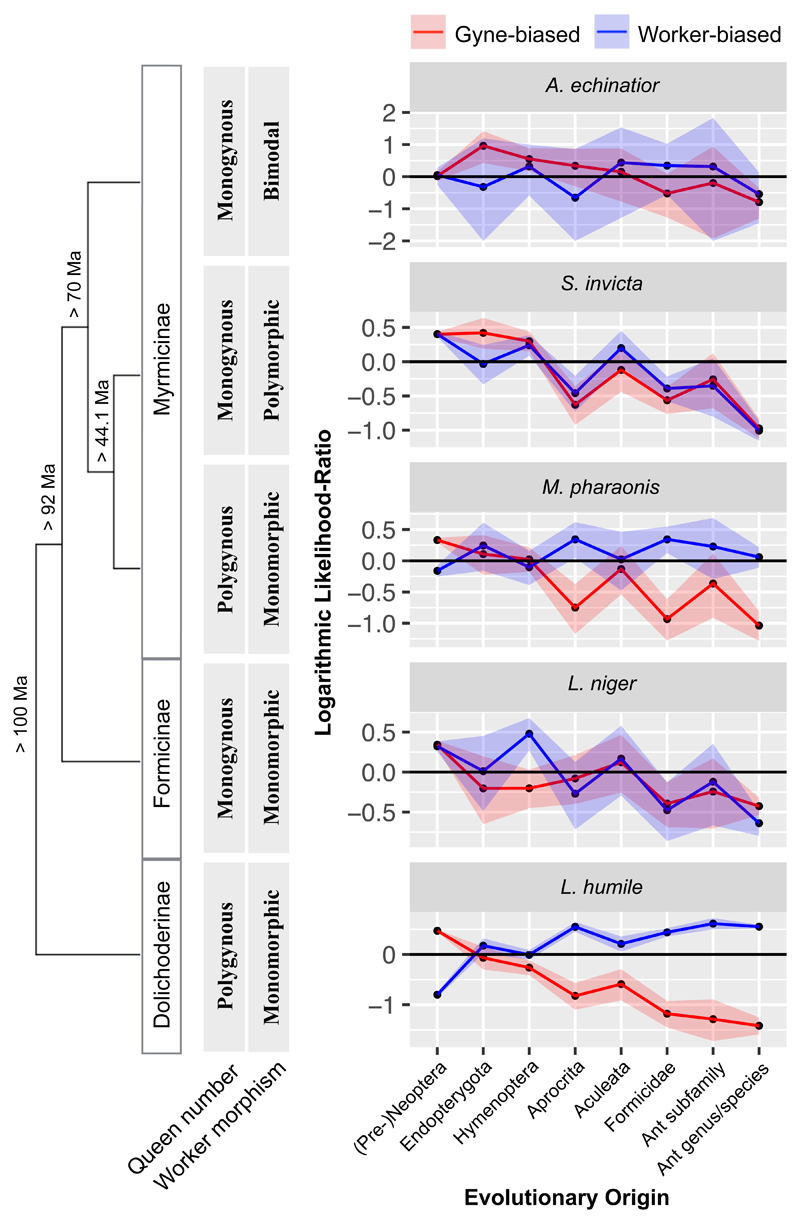
We produced and re-analysed brain transcriptomes for five ant species with normal caste differentiation: Acromyrmex echinatior, Monomorium pharaonis, Linepithema humile, Solenopsis invicta and Lasius niger 26, and for the queenless ants Ooceraea biroi 22 and Dinoponera quadriceps 27 which are, respectively clonal with egg-laying and sterile workers and sexual with secondary evolved dominance hierarchies where one worker becomes inseminated to serve as (gamergate) queen 28 (Supplementary Table 1). We recovered all orthologous genes across 23 re-annotated insect genomes and traced the evolutionary history of genes for all five species with normal caste differentiation (see Methods) to assign gene-origins in the Neopteran phylogenetic tree and later gene emergences in the ant tree. We then identified genes with caste-specific brain expression in each species and found that 42-62% of them had early Neopteran origins and 8-25% were ant-lineage specific, either at the subfamily or genus level. These percentages were similar but somewhat lower than those for the whole genomic background, which gave 45% (40-51%) genes with ancient homologs and 26% (18-35%) without (Supplementary Table 2).
We then used log likelihood-ratios (LR) to evaluate differences between proportions of caste-biased gene-origins at each node and proportions of extant genes with caste-biased brain expression, which allowed direct comparison between caste-biased genes of different evolutionary ages. Likelihood ratios of gyne-biased expression increased with evolutionary age in all five ants (all p < 1e-3; Figure 1), implying that early Neopteran genes are significantly more likely to have gyne-biased expression than genes of more recent origin. However, likelihood ratios of worker-biased genes varied, having similar frequencies as gyne-biased genes in A. echinatior (gynes versus small workers), S. invicta and L. niger, but slightly or steeply decreasing frequencies with age-of-gene-origin in M. pharaonis and L. humile, respectively (Figure 1). In the first three species, ancient genes had varying degrees of higher prevalence than lineage-specific genes (S. invicta: 10.9% versus 2.6%; L. niger: 4.9% versus 1.9%; both p < 1e-3. A. echinatior: 0.3% versus 0.1%, not significant: P = 0.29). However, in L. humile and M. pharaonis ancient genes were significantly less likely to have worker-biased expression than lineage-specific genes (L. humile: 10.4% versus 40.3%; M. pharaonis: 2.9% versus 3.7%; both P < 1e-3) (Figure 1) (Supplementary Table 3).
Figure 1. Likelihood-ratios of genes with caste-biased expression in the brains of five ant species originating at subsequent phylogenetic nodes.
All log2-likelihood-ratios are relative to the total number of genes with caste-biased expression in either direction, with positive and negative values indicating higher and lower likelihoods and shaded areas representing 90% confidence intervals based on binomial distributions. The ant subfamily tree on the left has been supplemented with key characteristics of typical social organization (monogynous = single-queen colonies; polygynous = multiple-queen colonies; monomorphic = unimodal size distribution of workers; polymorphic = skewed size distribution of workers as in S. invicta or bimodal distribution as in A. echinatior). For S. invicta we pooled equal amounts of large and small workers, so gene expression refers to average expression levels among different worker sizes. For A. echinatior we present likelihood ratios between gynes and minor workers here and those between gynes and major workers in Supplementary Figure 1. Estimated divergence dates are given along the tree, based on earliest fossil records and estimated phylogenetic divergence of ant clades clades 59,60.
The relative effects of species and caste identity on gene expression in ant brains
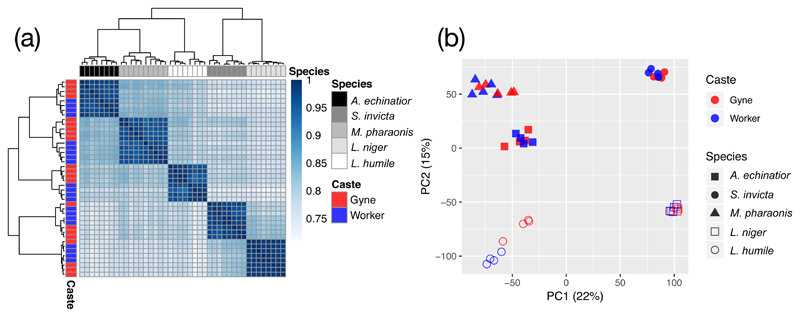
We identified 6672 orthologs that were shared across all seven ant species. Overall expression similarity matrices for the five ants with typical gyne-worker caste differentiation showed that brain samples from the same ant species clustered together regardless of caste (Figure 2a). Principal component analysis (PCA) of gene expression data clearly separated samples according to species identity, with the first two axes jointly explaining 37% of the total variance in gene expression (Figure 2b). However, within each species, samples of the same caste clustered more closely than samples from the same colony (Figure 2a) suggesting that brain gene expression patterns might predict gyne and worker phenotypes provided lineage-specific determinants of gene-expression could be removed.
Figure 2. Gene expression signatures of species identity and caste type, calculated from 6672 one-to-one orthologs across seven ant species.
(a) Expression similarity matrix for brain samples with each cell representing overall transcriptomic expression similarity between a pair of samples based on Spearman correlation coefficients across all orthologous genes, ranging from 0 (white; no correlation) to 1 (dark blue; perfect positive correlation). The type of caste represented by each sample is illustrated in the red/blue left vertical bar and species identities are given in the grey-scale horizontal bar above the plot. (b) The first two principal components explaining transcriptomic variation across brain samples with red/blue colours for castes and different symbols for species: closed symbols refer to the three species of the sub-family Myrmicinae and open symbols to the remaining two species belonging to the sub-family Formicinae (L. niger) and Dolichoderinae (L. humile). All 6672 one-to-one orthologs were used for both plots, and expression levels were normalized by log2 transformation of the number of Transcripts per Million kilobases (TPM) and then by quantile normalization across samples.
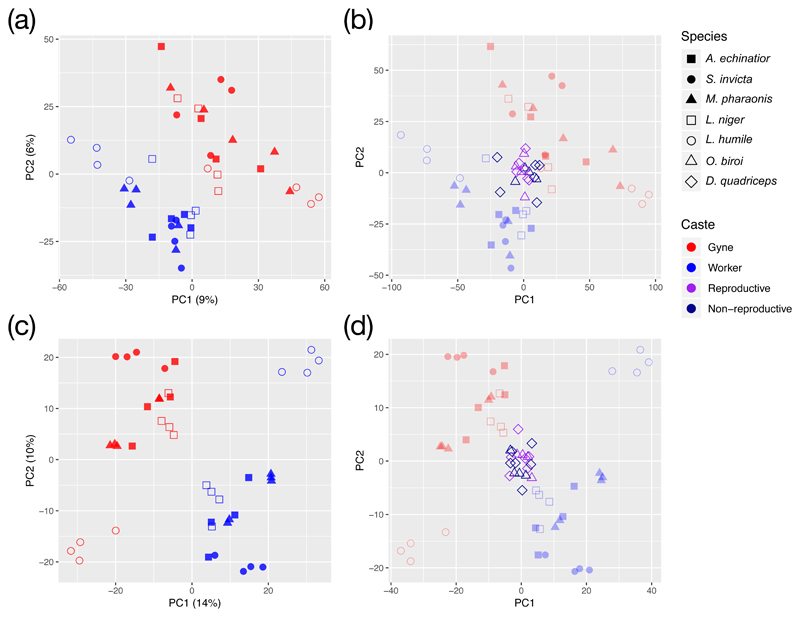
To separate effects of genus or subfamily identity and caste on brain transcriptomes we calculated species-specific z-scores by subtracting mean expression levels of each gene and normalizing also for variation in gene expression across samples. This produced a matrix of directly comparable residuals across species, which showed that brains of same-caste individuals have similar patterns of gene expression (Supplementary Figure 2a). The first two PC-axes explained 15% of the overall variance and completely separated all samples according to caste (Figure 3a). Similar patterns were found when transcriptome data were pre-processed with other methods such as regularized log transformation (rlog) or variance stabilizing transformation (VST) (Supplementary Figure 3) 29. A coherent signature of caste-specific gene expression in ant brains thus appears to exist, even though we could not adjust for epistatic and environmental confounders that are likely to also be important. Normalizing for colony-level effects further improved the overall resolution for caste-specific gene expression with the percentage of variation explained by the two PC-axes increasing to 24% (Figure 3c and Supplementary Figure 2b) (Methods).
Figure 3. PCAs for brain transcriptomes calculated from 6672 one-to-one orthologs across seven ant species, after adjusting for either species-level or colony-level variation in the mean and variance of gene expression.
PCAs for brain transcriptomes after partialling out additive effects of species identity (a) and species plus colony identity (c) in the same five ant species as in Figures 1 and 2. Projection of similarly normalized brain transcriptome scores for the queenless ants O. biroi and D. quadriceps (b and d; foreground darker symbols) on the two PCs of (a) and (c). All 6672 one-to-one orthologs were used in both plots and colour coding and symbols are identical to Figure 2b for the ant species included in both analyses. Additional symbols and colours were used to differentiate between reproductive and non-reproductive workers of the two queenless ants.
We used 2-way ANOVA to unravel confounding effects of ant subfamily (Figure 1) using the scores along the first two principal axes describing transcriptome variation after species-level normalization. This showed that caste remains the dominant predictor along both PC-axes, explaining 44.5% and 42.8% of the respective variances (P = 8e-9 and 6e-8) (Figure 3a). Sub-family did not appear to have a directly significant effect (explained variance = 1e-4 for PC1 and 1e-5 for PC2; both P = 0.99), but 26.5% and 23.4% of the respective variances along the 1st and 2nd PC-axes were explained by the interaction term between caste and subfamily identity (P = 8e-6 and 7e-5). Using colony-level normalization produced a similar result (Supplementary Table 4), suggesting that future comparisons across a phylogenetically more diverse set of ants may well reveal clear sub-family effects on brain gene expression across castes in addition to the genus/species-level effects that we document.
Brain transcriptomes in ants that secondarily gained a worker caste or lost the queen caste
Additional worker and soldier castes have arisen in many ant lineages, normally with body sizes intermediate between queens and small workers 30–32. Within the attine ants, which evolved 55-60 million years ago (Ma), only the evolutionarily derived Atta and Acromyrmex leaf-cutting ants that originated ca. 15 Ma 33 have such additional castes as secondary innovations. We compared brain transcriptome similarity (normalizing again for species identity) of large and small A. echinatior workers relative to workers of the other four species. This showed that large-worker brain transcriptomes were distinct from those of gynes but that their expression profiles were more distant from those of the monomorphic workers of the four other ant species (Supplementary Figure 4), as expected when large-workers are evolutionary derived. Also these resolutions further improved when we normalized for colony-level variation (Supplementary Figure 4).
The queen caste has secondarily disappeared in several ant lineages. This happened before workers had irreversibly lost the sperm storage organ (spermatheca) so mated-workers could evolve to be ergonomically cheaper egg-layers when colonies were small 34,35. Rarely, ant lineages abandoned sexual reproduction altogether 36,37 to produce new workers from unfertilized worker eggs 22,27. Using published brain transcriptomes of reproductive and non-reproductive workers (Supplementary Table 1), we found (after species-level normalization) that clonal reproductive and non-reproductive O. biroi workers were separated by the 1st PC axis and that separation once more improved after normalizing for colony identity. Brain gene expression differences were less pronounced in D. quadriceps where colony-level normalization was needed to make the two worker phenotypes segregate in most but not all colonies (Supplementary Figure 5a). In both species the expression patterns of fertile and sterile workers segregated along the same PC axis, suggesting there may have been convergent evolutionary responses to losing the queen caste (Supplementary Figure 5b).
We used singular-value decomposition (SVD) to assess overall gene-expression similarity between the two queenless and five other ants. We first validated this method for the normalized brain transcriptome data of the five ants via leave-one-out jack-knife resampling, which assigned four ants as training data and then tested the match between expected and observed caste-specific expression in the fifth species. This showed that segregated gene-expression profiles of queens and workers could be retrieved for all five combinations, and again more consistently after normalizing for colony-level variation (Supplementary Figure 6). Projecting the queenless ant data onto the five normal ant species PCA matrix showed that brain transcriptomes of their reproductive and non-reproductive workers fell in the centre of the plot and failed to segregate, both after normalizing for species-level and colony-level differences (Figure 3b and Figure 3d). With the first two PC axes likely representing the ancestral GRN for differential queen-worker gene expression, this result is consistent with both queenless ants having lost the ancestral caste GRN (Supplementary Figure 5) although the genes composing the GRN are still present. We further tested whether phenotypic plasticity for reproductive roles within a single female bauplan would generally fail to produce segregation along our PC-axes for genetically hard-wired castes by plotting brain gene-expression data for breeders and helpers of Polistes canadensis wasps 27. This showed a similar lack of segregation, consistent with these wasps lacking permanent castes because they never passed the point of no return to superorganismality 3 (Supplementary Figure 7).
Key ancestral genes in the genetic regulatory network mediating queen-worker caste differentiation in ants
To elucidate functional categories of gene-sets regulating gyne-worker caste differentiation, we identified pathways with consistent caste-biased expression across the five ant species using enrichment analysis 38. After further colony-level normalization we found that metabolism-associated genes (e.g. oxidative phosphorylation and carbon metabolism) were significantly up-regulated in gynes across the five ant species. Also the photo-transduction pathway was up-regulated in gynes except for L. niger where a same-direction difference was not significant. Such enrichments were not detected in the reproductive workers of both queenless ants, as expected when they lack specialized queen phenotypes that need vision during mating flights (Supplementary Table 5, Supplementary Figure 8).
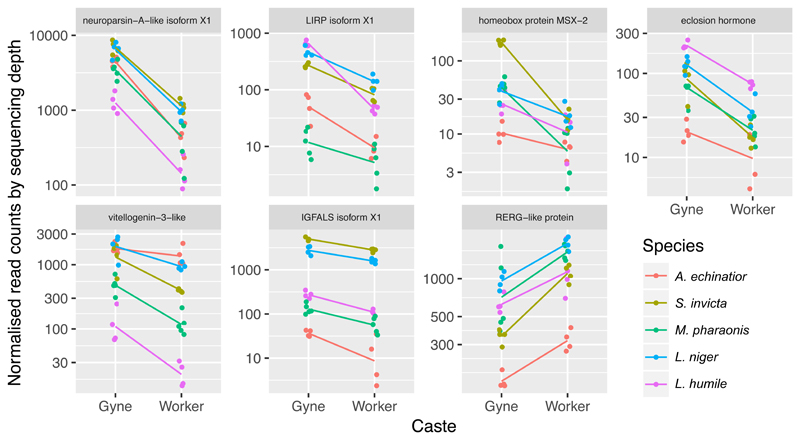
Using a generalized linear model (GLM) to account for additive effects of caste and species identity on expression of the 6672 orthologous genes, we identified 42 genes with significant differential expression between gynes and workers across all five ant species (38 up-regulated in gynes, 4 up-regulated in workers; fold change > 1.5; FDR < 0.01). We interpreted these as potential key genes in the conserved ancestral GRN for queen-worker differentiation (Supplementary Table 6). Consistent with the gene-set/pathway enrichment results, several of these genes are involved in vision, including neither inactivation nor afterpotential protein C (ninaC) and the protein bride of sevenless (boss). Also genes associated with hormonal and insulin systems, including neuroparsin-A like, vitellogenin-3-like, eclosion hormone, locusta insulin-related peptide (LIRP), and insulin-like growth factor-binding protein complex acid labile subunit (IGFALS), were significantly up-regulated in gynes across the five ant species (Figure 4), whereas Ras-related and estrogen-regulated growth inhibitor-like protein (RERGL) was significantly up-regulated in workers (Figure 4).
Figure 4. Expression levels for conserved caste regulatory genes expressed in the brains of gynes and workers across the five ant species with typical queen-worker differentiation.
Dots are gene expression levels for each brain sample and lines connect the average brain gene expression values for gynes and workers (both coloured according to species identity; See text for details). All expression differences between gynes and workers were significant (FDR < 0.01, two-sided Wald tests).
To tentatively explore the identity of the ancestral ant GRN components, we constructed a cross-species gene co-expression matrix (see Methods), which produced clusters of genes exhibiting conserved co-expression with the key genes of the conserved ancestral GRN that were nested within these clusters (Supplementary Information). These networks seemed to consistent with the extant ant GRN having maintained overall aspects of co-regulation, but this result will need validation and refinement with species-specific transcriptome data for time-series of developmental phenotypes.
Toolkit genes involved in convergent caste differentiation in ants and corbiculate bees
The corbiculate bees (honeybees, stringless bees, bumblebees) and the ants independently evolved the permanent castes 39 that define superorganismality 3. It has been hypothesized that there exists a toolkit of genes that are differentially expressed in breeder and helper phenotypes across insect lineages 9,40, but whether such genes play analogous roles in independent transitions to superorganismality remains unknown. We therefore compared the underlying GRNs for caste differentiation between ants and honeybees using pupal honeybee brain transcriptome data of orthologous genes 41. Among the 42 conserved caste-GRN genes in ants, 39 have orthologs in the honey bee and 15 of these (38.5%) had significant differential expression between honeybee queens and workers (fold change > 1.5; P < 1e-3; Supplementary Information). This percentage is higher than the overall proportion of genes with any differential expression across honeybee gynes and workers (733/6036 = 12.1%) (P < 1e-4), consistent with an ancestral hymenopteran toolkit of genes mediating reproductive division of labor 9. However, only five of these 15 genes had the same direction of caste-biased expression between ants and honeybees (an uncharacterized protein LOC105150705, elongation of very long chain fatty acids protein AAEL008004-like, ninaC, neuroparsin-A-like, and homeobox protein MSX-2; Supplementary Table 6), a ratio not significantly different from the 12.1% overall proportion (P = 0.58). This lack of directional consistency in caste-biased expression implies that the ancestral GRNs of ants and honeybee are uncorrelated in spite of including some of the same toolkit genes, as expected when irreversible transitions to superorganismality evolved convergently.
Independent recruitment of genes with caste-biased expression was confirmed when we considered all orthologous genes between ants and the honeybee. After normalization ant gynes and workers segregated along the first PC-axis and honeybee gynes and workers along the second PC-axis (Supplementary Figure 9a). When we projected the honeybee caste-specific brain transcriptomes on the first two PC-axes for ant transcriptomes, the second PC-axis separated gyne and worker brain samples of the honeybee, but with expression bias in opposite direction compared to ants (Supplementary Figure 9b).
Discussion
Our study reconstructs the contours of the ancestral brain GRN in queens and workers 20,42 that established permanent reproductive division of labor in the ants 43,44. We found that later subfamily/genus effects blur this conserved ancestral GRN 23, as expected after ca. 120 Ma adaptive radiation 20,43. This finding underlines the necessity of elaborate normalization procedures, which possibly explains why our study, based on a single matrix approach across species, achieved higher resolution than Morandin et al. 8 (Supplementary Information). Some ant GRN genes, such as vitellogenin-3-like and neuroparsin-A like, are homologs of juvenile hormone genes 40,45 involved in regulation of facultative and obligate reproductive division of labor across social Hymenoptera 40,46,47. However, the myosin light chain gene, reported as differentially expressed between castes across 16 ant species 8 was not part of our GRN. This gene was slightly over-expressed in gynes (rather than under-expressed as in 8) in four ant species (not in L. niger), suggesting it exemplifies the problematic resolution of whole body transcriptomes, because it must have had opposite caste-biased expression in other body parts. This previous study 8 used overlapping species comparisons to identify common genes with caste-biased expression, which implied that type-2 errors increased proportionally. Our GLM approach avoids this problem by integrating all cross-species expression levels into a single data matrix after multiple normalization procedures, so that sampling variance in the number of detected ancestral DEGs becomes gradually less when more species are included in a comparative analysis (Supplementary Information). Three quarters of the genes with caste-biased expression across the five ants that our study discovered have not been identified in this role before, corroborating the improved resolution that our methodological approach offers for future comparative transcriptomics studies of social evolution questions.
Our results support the notion that permanent reproductive division of labor between specialized queens and life-time unmated workers was a major evolutionary transition in the ancestral ants 1,3, involving a single genetically hardwired point-of-no-return that established the brain GRN for caste. Our analyses beyond the five ants with normal caste differentiation are consistent with genetically hardwired caste differentiation being fundamentally different from what phenotypic plasticity can achieve, both in ants that secondarily lost the queen-caste and in dominance hierarchies of cooperatively breeding Polistes paper wasps that never evolved permanent castes. As envisaged by Wheeler (1911), only superorganismality based on permanently differentiated castes invites analogies with metazoan multicellular development. One of these is serial homology, as we found for the brain GRNs of the two distinct worker castes of A. echinatior, a pattern functionally reminiscent to transcriptomes specifying the upper and lower first molars of mice that originated from a single tooth 48. Our results also confirm that the point-of-no-return transition to superorganismality in the corbiculate bee lineage (bumblebees, stingless bees, honeybees) 39,49 was independent from the ants. Our study predicts that brain GRNs for permanent castes between these three corbiculate bee lineages should be mutually correlated because they share a recent common ancestor, but uncorrelated with the queen and worker brain GRN in vespine wasps, which evolved superorganismality independently from ants and bees. Larger-scale brain GRN studies across the superorganismal lineages as defined by Wheeler 1,3 will thus be illuminating to further explore lineage-specific elaborations of social organization, such as the tentative link that we report between recruitment of novel genes for worker functions and obligate polygyny in ants (Figure 1; Supplementary Information).
Independence of gene-expression directionality in ants and the honeybee does not preclude that orthologous genes are part of ancestral genetic toolkits across convergently evolved superorganismal lineages 9. The toolkit concept is about participation as such 9,23, whereas the ancestral GRN concept is about correlated co-expression across castes. The existence of the former is purely an empirical issue, but the latter has predictive elements because co-expression should be similar across lineages derived from the same major transition to superorganismality, but not across independent irreversible transitions of this kind 3. Our findings were consistent with this expectation, but the details of this inference remain somewhat provisional because transcriptomes were from adult brains in ants and from pupae in the honeybee. Although we partly validated these results by comparing pupal transcriptomes from the honeybee and the fire ant (see Methods), direct comparison with transcriptomes of virgin adult honeybee queens are needed. More comparative brain transcriptome studies of ancestral GRNs across the ant, bee and wasp lineages with permanently unmated workers will undoubtedly add to our understanding of the selection forces that convergently produced analogous forms of superorganismal social organization.
Methods
Code availability
The Python and R scripts used to process the data are available on https://github.com/StanQiu/ant_brain_comparative
Sample collection
Acromyrmex echinatior
Five colonies of A. echinatior were collected in Panama in 2004 (Ae263), 2014 (Ae704) and 2016 (Ae747, Ae764, Ae767) and reared at the Centre for Social Evolution, University of Copenhagen under a constant temperature of ca. 25°C and ca. 70% humidity. Gynes and minor workers were collected within the fungus garden, whereas major workers were collected while they were foraging outside, always during daytime (10 am to 4 pm), and isolated in groups of 6-10 individuals of the same caste in small fluon-coated plastic boxes. Ants were then put on ice to reduce activity before their brains were dissected in a dissection-dish with Diethyl pyrocarbonate (DEPC) treated Phosphate-buffered saline (PBS) on ice. Dissection was done under a stereomicroscope with sterile (heat-treated) forceps and only samples consisting of the complete and undamaged anatomical structure of a brain were retained. Each brain dissection was completed within five minutes and dissected brains were transferred to 1 ml RNAlater (pooling maximally 12 brain samples of the same caste), kept at 4°C overnight, and then transferred to a -20°C freezer for long term storage.
Monomorium pharaonis
Five colonies of M. pharaonis (3rd Room X1/B, CS10, Donor 3rd, Donor BQ-, 3rd Room X1/A) were reared at the Centre for Social Evolution, University of Copenhagen under constant temperature of 27°C and 50% humidity from a stock collected in 2008 50. Gynes were separated from males at the pupal stage and reared with 10–15 workers in fluon-coated petri-dishes to be collected within three days after they hatched as adults, whereas workers were collected directly from the source colonies when they were foraging. All collections were done during daytime (10 am to 4 pm) and adults were kept in groups of 6-10 of the same caste in small fluon-coated boxes. Dissection procedures were the same as in A. echinatior, except that dissected brains were transferred into RNAlater with a droplet of PBS because brains were too small to be handled directly with forceps. Upon storage, each 1ml RNAlater sample contained at most six brain samples of the same caste to ensure RNAlater concentration remained high enough.
Linepithema humile
Five colonies of L. humile were collected in Caldes d'Estrac, Spain (Catalan3b) and Castell d'Aro, Spain (Main 3a, Main 4a, Main 5b, Main 5d) in 2016 and reared at the Centre for Social Evolution, University of Copenhagen under constant temperature of 27°C and 50% humidity. Gyne isolation, worker ant sample collection, brain dissection, and storage proceeded in the same way as for Monomorium pharaonis.
Solenopsis invicta
Four monogynous (single queen) colonies were collected in Taoyuan, Taiwan, two in October 2012 and two in April 2014, and transferred to a fluon-coated plastic box in the lab. Approximately 120 gynes and 200 workers from each colony were randomly selected for dissection. For workers, an equal number of small and large workers were selected. Ants were cooled on ice, then soaked into RNAlater and dissected with forceps in RNAlater on ice. Forceps were dipped into ethanol and flame sterilized between samples. Each dissection was completed within five minutes and then transferred into tubes with 50 ul Trizol (maximum 12 brain samples of same castes per tube) and stored at -80°C. All sampling and dissections were done within one month after collection in the field.
RNA extraction, cDNA library construction and RNA sequencing
A. echinatior, M. pharaonis and L. humile brain samples were retrieved from their RNAlater storage tubes after which 10 - 12 brain samples for each caste and colony were pooled before RNA extraction. Total RNA was extracted using the QIAGEN RNeasy Plus Micro Kit (cat. no. 74034) according to the manufacturer’s protocol (2013), and quality was evaluated with an Experion HighSens RNA analysis assay (Bio-Rad) prior to library preparation. Total amounts of RNA were quantified using an Invitrogen™ Qubit RNA High Sensitivity assay. Based on the RNA sample concentration, Ambion® ERCC RNA Spike-In Mix (cat. no. 4456740) was added to each sample according to the manufacturer’s instructions. cDNA libraries were then constructed using a Clontech SMARTer PCR cDNA Synthesis Kit (cat. no. 634925), following the protocol described in the manufacturer’s user manual (Protocol No. PT4097-1). The numbers of PCR cycles for the 2nd strand cDNA synthesis reactions were optimized as described in the user manual. RNA sequencing was then conducted at BGI’s Sequencing Centre, Wuhan on an Illumina HiSeq4000 platform with 150 bp paired ends reads. We have generated around 3-5 Gbp of RNAseq data for each sample (Supplementary Table 1).
For S. invicta, brain samples (already in Trizol) of the same caste and colony were first pooled and then homogenized with ceramic beads in a bead shaker (MP Fastprep-24), after which chloroform was added to separate the liquid phase according to a standard Trizol protocol. Afterwards, the aqueous phase containing the total RNA was directly applied to the GE Illustra RNAspin Midi kit for RNA purification according to the manufacturer’s protocol. RNA sequencing was conducted at Academia Sinica High Throughput Genomics Core, Taipei, Taiwan. All samples were sequenced on an Illumina HiSeq2500 platform with 101 bp paired ends reads. For one set of biological replicates, additional sequencing was obtained on an Illumina GA platform with 96 bp paired end reads and an Illumina MiSeq platform with 250 bp paired end reads. This generated 4-10 Gbp of RNAseq data for each sample of S. invicta (Supplementary Table 1).
We further included published brain transcriptome data for Lasius niger 26, Ooceraea biroi 22, and Dinoponera quadriceps 27 in our comparative analyses, each with four to six colony replicates (see Sample collection and Supplementary Table 1).
Quality checks of the RNAseq data
For the three sets of RNA samples with ERCC-spike-in, data quality was checked with mapped reads (log2 transformed and with one pseudo-read to avoid log2(0)), so we could remove outlier samples if their spike-in Pearson correlation coefficients across samples were < 0.95. Similarities in caste-biased gene expression were also used to check data quality: For each ant species, mapped transcriptome reads (log2 transformed as mentioned above) were compared among samples within the same caste of a focal species and checked for potential biological or technical biases when producing within-caste Pearson correlation coefficients < 0.9, as suggested by the ENCODE RNAseq guidelines 51. These checks implied that we excluded seven outlier samples collected from deviating locations (assuming biological reasons for being outliers) or being prepared with somewhat deviating experimental procedures (assuming technical reasons for being outliers). If no associations with potential biological or technical biases could be identified from our notebooks, samples were retained for downstream analysis.
Improved genome annotation for all seven ant species based on RNAseq data
The genomes of the seven ant species included in this study, A. echinatior, M. pharaonis, L. humile, S. invicta, L. niger, D. quadriceps and O. biroi, were re-annotated based on the NCBI genome annotations of all these seven species plus Drosophila melanogaster and Nasonia vitripennis. The re-annotations were done with GeMoMa 52, a method based on the assumption that amino acid and intron positions are conserved across the insect phylogeny. This tool was chosen as particularly suitable for our project because the explicit use of exon-intron-structure information was shown to improve annotation accuracy 52, and because integrating genome annotations across ant species increases the discovery rate of orthologous genes in each ant lineage. For each target ant species, the genome was first re-annotated by independent comparison with the genome annotations of the other ant species and the two non-social insects, and RNAseq data for the target ant species were then used to pursue re-annotation for each of the ant genomes. The re-annotated genomes of the target ant species were finally filtered for the combined analysis, using the genome annotation filter (GAF) provided by GeMoMa, to remove redundant or overlapping exons before they were used as reference genomes for RNAseq analysis. Overall, these re-annotations increased the total number of annotated genes across all ant genomes by 27% (from 90,856 to 115,739), which increased the accuracy of our orthology inferences.
Improved genome annotations for 16 other social and non-social insect species
To infer the origins of orthologous genes in the most accurate way possible, we also re-annotated the genomes of Harpegnathos saltator, Apis mellifera, Bombus terrestris, Habropoda laboriosa, Melipona quadrifasciata, Trichogramma pretiosum, Copidosoma floridanum, Ceratosolen solmsi, N. vitripennis, Orussus abietinus, D. melanogaster, Mochlonyx cinctipes, Tribolium castaneum, Pediculus humanus, Frankliniella occidentalis, and Zootermopsis nevadensis (Supplementary Figure 10). For each of these insect species, we used the six improved ant genomes (excluding L. niger because we added this species at a later stage of analysis) as references and re-annotated the insect genomes using GeMoMa as above, except for the integration of RNAseq data as they were only available for the seven-ant species.
Finding gene-orthologs across the 23 insect genomes
Orthologous genes of the seven-ant species were identified in four steps. For the orthologs among the 16 re-annotated insect genomes other than the seven focal ant species of our study we: 1) retained the longest protein sequence for each gene when we obtained more than one transcript, and 2) performed reciprocal best hit BLASTP with an E-value < 1e-5 and sequence coverage >50%. For the seven ants, we then, 3) used the homologs that both fell within the 95% confidence interval of being orthologs in reciprocal BLASTP searches, and confirmed assignments by additional synteny information because only orthologs should have remained co-linear with at least their immediately neighbouring genes, and 4) retained only genes with exactly one ortholog for each of the seven ant species so that inter-specific transcriptome comparisons were always based on one-to-one ortholog matches. Steps 2 and 3 were done using Proteinortho 5.16 with default settings 53.
Determining the origins of orthologous genes
Using the package Proteinortho 53, the origin of each gene was determined by integrating the ortholog-information and the phylogenetic relationships among species (Supplementary Figure 10) using three criteria. First, if a reference gene had one or more orthologs in a branch with multiple species, we considered that to be sufficient evidence for the occurrence of a target gene’s ortholog in a branch. Second, the occurrence of a target gene’s orthologs along phylogenetic branches was conceptualized as a binomial process, and the probability of missing an ortholog (i.e. the expected rate of loss of a gene) on a branch was set to be 10%. This threshold is somewhat arbitrary, but allowed missing orthologs (either due to gene loss or miss-annotation) in recent branches to have somewhat higher probability while still considering ortholog losses to be independent among branches. This procedure thus controlled for the number of missing orthologs along branches, i.e., for every candidate ortholog occurring in the earliest branch, we examined the inferred orthologs distribution along later branches and discarded a distribution as pseudo-orthologous if the probability was < 0.01, after which we continued the same procedure for the next older branch. This refers to situations where orthologs occurred in an ancient phylogenetic node without finding corresponding orthologs in multiple recent lineages. Third, the origin of a focal gene was considered to be unambiguously identified with the oldest paired ortholog occurrence in the phylogenetic tree.
To illustrate the robustness of our approach we present results for a 10% rate of gene loss in the main manuscript and provide complementary analyses for putative rates of 5% and 20% in Supplementary Figure 11.
Likelihood estimation of the nodes where caste-biased gene-expression evolved
The likelihood (LR) of caste-biased gene expression at each phylogenetic node (i) was calculated as the log ratio between the proportion of genes with specific caste-biased expression in a focal evolutionary node (n) relative to the overall proportion of genes with the same type of caste-biased expression in the transcriptome (N),
| (1) |
where caste can either be gynes or (subcategories of) workers. The ratio of n(i)caste / n(i) and Ncaste/ N also produced a confidence interval of the LR estimates when assuming that recruitment of genes for caste-biased expression follows a binomial distribution.
Transcriptome quantification
Transcriptome quantifications were done with the Salmon pipeline 54. In brief, RNAseq data for the seven-ant species were quasi-mapped to the corresponding genome with improved annotation (as described above), after which bias-correction options were turned on to account for GC-bias and sequence-specific bias. For the RNAseq data of A. echinatior, M. pharaonis, and L. humile, where ERCC-spike-in was added before cDNA library construction, additional quantifications were done with corresponding genome + ERCC pseudo-scaffolds.
Identification of genes that are differentially expressed among castes
For each ant species, genes with differential expression among castes were identified using the RNAseq data from the samples that had passed quality checking. Analyses were done with the DESeq2 package in R 29 as follows: (1) We used transcript-level read counts from Salmon as input for DESeq2, (2) We aggregated the transcript-level read counts to become gene-level read counts using tximport, (3) We modelled the gene-level read counts as: count ~ caste + colony, assuming that the influence of caste and colony on gene expression levels is additive; this enabled us to include the possibly confounding effects of colony origin on caste specific gene expression levels of colony members, and (4) We tested for gene expression differences between castes using DESeq2 and adjusted P-values after independent hypothesis filtering (IHF), which controls for false discovery due to multiple testing while increasing detection power 55. Genes were identified as being differentially expressed between castes when the adjusted P-value was < 0.05 based on a two-sided Wald test.
For the honey bee data in our subsequent comparison with ant data, we used recently published brain transcriptome data of the P4 stage of worker pupae and the P3 stage of gyne pupae, as these two stages represent the same developmental age 41. We used the same procedure to detect differentially expressed genes between castes as in our ant comparisons, except for using a more stringent fold-change expression difference between castes as threshold for acceptance of DEGs, i.e. we kept 1.5 fold-change expression difference between castes as null model and used a more stringent P-value < 1e-3 instead of 1e-2 used in our ant species comparisons. See Supplementary Information for a comparative evaluation of P-value cut-offs.
While the choice of P-value or fold-change cut-off will affect the number of genes identified as differentially expressed (detection power), other technical and experimental design differences will also affect detection power, which makes it difficult to remain fully consistent when using data from multiple sources as we do in the present study. However, as our PCA and clustering analyses used whole transcriptomes (i.e., the expression level of all genes in brains), we did not rely on specific P-values or fold-change cut-offs for our overall comparisons of caste specific transcriptomes among ant and other social insect species. This implies that our main conclusions (of consistent directional gene expression differences in queens versus workers across ant species, and of genetic regulatory networks between ants and the honeybee being uncorrelated in their direction of expression bias) are independent of the choice of P-value and fold-change cut-off.
We used pupal brain transcriptome data of honeybee queens and workers for comparison with ant transcriptomes because adult brain transcriptomes (RNA-seq data) were not available. Using only adult caste data would have been preferable, but we partially validated our comparisons by repeating our analyses (data not included) using pupal brain transcriptome data of gynes and workers in S. invicta, which showed they segregated for the same two PC axes as the brain transcriptomes of adult gyne and worker castes in the five ant species. Also direct pupal comparisons between S. invicta and the honeybee recovered the distinctly opposite patterns of directionality in gene expression that we report. We therefore assume that the conclusion of our comparisons of gene-expression bias in ants and the honeybee are reasonably robust, but we have added a caveat on this in the Discussion of the main paper.
Normalization of gene expression scores across samples
To compare transcriptomes across samples, expression levels for genes with one-to-one orthologs in all seven-ant species (n = 6672) were normalized across samples. Normalizations were done as follows: 1) For each sample, Transcripts per Million Kilobase (TPM) for each gene were increased with 1e-5 pseudo transcripts (to avoid log(0) scores) and then log2 transformed, 2) The log2 transformed transcriptome data were then quantile normalized among samples, i.e., by ranking the expression levels for all genes in each sample and then replacing gene-specific expression levels with rank numbers across samples to equalize the average expression level scores. This quantile normalization ensured that the overall distributions of gene expression levels remained the same across samples so that effects of any technical artefacts were minimized 56.
KEGG pathway analyses and Gene Ontology (GO) analyses
Gene Ontology and KEGG annotations for ant genomes were done by comparing homologous annotations between ants and D. melanogaster and integrating these types of information in three steps. First, genes in the ant genomes were compared by BLASTP to those in Drosophila melanogaster. Second, gene-pair values with BLAST E values < 1e-5 and query coverage >30% were retained. Third, ant genes were annotated with Entrez ID from their fruit fly homologs and linked with EntrezID values from the Gene Ontology and KEGG annotations.
To find gene-sets with the most consistent caste differences in gene expression across ant species, we examined brain gene expression differences between gyne and worker samples across colony replicates in the five ant species with typical caste differentiation: A. echinatior, S. invicta, M. pharaonis, L. niger and L. humile. We omitted O. biori and D. quadriceps from this analysis because they lack the queen caste and have secondarily evolved egg laying by parthenogenetic or inseminated workers (gamergates), respectively. We used the quantile normalized expression values of the 6672 orthologous genes (See section: Normalization of gene expression scores across samples above) as input for gene-set enrichment analyses using the General Application Gene-set Enrichment (GAGE) package in R 38. Colony-specific gyne samples were always marked as ‘target’ and worker samples as ‘reference’, and we fixed the “compare” option as Paired when comparing differences in gene expression between gyne-worker pairs for colony replicates. Enrichment analyses were done using KEGG and Gene Ontology (GO) annotations from corresponding homologs in the fruit fly. We fixed the “same expression direction” option as True in both KEGG and GO enrichment analyses, which allowed us to detect gene-sets that are consistently up or down regulated in gynes relative to workers. Gene-sets were identified as being significantly enriched when false discovery rates (FDR) were < 1e-2 in two-sided non-parametric Kolmogorov-Smirnov tests.
Adjusting for species and colony identity to obtain a general assessment of caste-specific gene expression in ants
A transcriptome with a total number of n genes in any sample can be regarded as an n dimensional vector , where = (X1, X2, …, Xn) and Xi represents the expression level of gene i in a particular sample. This expression level Xi is then affected both by its species-specific background (e.g. caused by species-specific developmental constraints, adaptations, or environmental conditions) and its actual caste state that should require the phenotypic expression of specific morphological and/or physiological traits. It is useful, therefore, to decompose vector as: where represents the influence of lineage(species)-specific genetic background on gene expression, represents the influence of the actual caste state, and represents any other effects on gene expression that are sample-specific, such as environmentally induced colony-level difference or any technical biases that might have affected single samples. To compare across multiple species, it is then essential to subtract and from . Assuming the effect of colony is small compared to the effect of caste and because samples of the same species were always prepared with the same experimental procedures, can be assumed to be the same within species and thus to group together with in a new vector . Thus, the task then becomes to obtain an estimate of for each species that can be used to partial out any confounding effects on caste-biased gene expression. Because Ci, the influence of caste phenotype on the expression of gene i in a particular sample, is either negative or positive and we expect a bimodal distribution of these values, we can assume that the distribution of Ci across samples of the same species is symmetrical around zero, also because we collected equal numbers of samples of different caste phenotypes. The sum of the Ci scores across multiple samples of the same ant species will then be 0 and the same applies to the sum of . Then the sum of across multiple samples of the same species can be expressed as: sum() = sum() + sum() = sum() + 0, so the average() = . This implies that = – = – average(), so that it is legitimate to compare across different species.
Based on this principle, we subtracted the mean expression levels within species for each gene to obtain quantile normalized orthologous transcriptome data controlled for species-level differences in the overall levels of gene expression. This was done with the SVA package in R 57, using the Combat function by setting species identity as a batch covariate and setting the mean.only option as False to also correct for differences in the total ranges of gene expression which are affected by absolute expression level difference across species. This normalization method thus removed all interspecies differences in gene expression profiles, so the mean and variance of expression levels for each gene became the same across species. The remaining residuals then reflected sample (colony-level) differences within species so that samples with similar residuals now represented similar caste-biased gene-expression values across species.
One of the key assumptions in the procedure outlined above is that the effect of colony of origin on brain gene expression is small compared to the effect of caste phenotype. However, because comparisons in the two queenless ants were among plastic phenotypes (egg-laying or not) within a single morphological caste rather than between two permanently differentiated castes, the colony-level variation was expected to be relatively more important. Thus, in these species we subtracted mean colony-expression levels while also adjusting for unequal variation in gene expression for each gene to obtain quantile normalized orthologous transcriptome data controlled for colony-level differences in overall gene expression. However, this made us realize that comparison with the five other ant species that had both caste phenotypes should ideally be based on samples that had undergone the same normalization procedures. We therefore always present results after species-level and colony-level normalization side-by-side. The colony-level normalizations normally explained higher proportions of the variance, but this was due to each colony only having one sample for pooled gynes and one for pooled workers, whereas species-level normalizations had 5 colony-level replicates. The higher percentages of explained variance after colony-level normalization therefore appear to mostly reflect a reduction in the error-term and should therefore carry similar but not quite identical weight as the species-level normalized results. This is because species-level normalization also removes any technical noise across pooled samples of 10 individuals, due to e.g. RNA extraction, PCR cycles and sequencing methodology, but variation of this kind should have remained very minor.
Constructing similarity matrices across samples, castes and species
Sample similarity matrices were calculated using 1- rs (Spearman correlation coefficients) as measure of gene expression distance between samples after quantile normalization procedures. Overall caste-specific expression similarity matrices were thus constructed according to the principles outlined above, using all orthologous gene data and the hierarchical clustering (hclust) function in R.
ANOVA to examine variation along the first two principal component axes for caste-specific gene expression
We used PCA to evaluate the similarity of genome-wide caste-specific gene expression patterns across the different ant species. Because each PCA axis obtained is a composite of many gene-level expression values, we examined the distribution of these scores using 2-way ANOVAs with sub-family identity and caste state as main factors and their interaction term, giving the model:
This analysis allowed us to estimate the proportion of variance in PC scores that could be explained by sub-family identity, a component that should be zero after adjusting for species level differences. The other main factor, caste state (morphologically differentiated gynes or workers) was also categorical, and the statistical interaction between these two main effects can thus be understood as a sub-family-specific overall effect on observed patterns of caste-biased gene expression.
Identifying caste-biased differentially expressed genes across the five ant species
Genes that are differentially expressed between gynes and workers across the five ant species were identified with a generalized linear model (GLM) in DESeq2 29. RNA read counts for each sample were imported after which we estimated the library size for each sample within the same ant species to adjust for library size differences, and subsequently modelled the expression levels of the 6672 orthologous genes as:
This assumes that the effect of species identity and caste state are additive, and will thus identify genes with consistent caste biased expression across the five ant species with normal caste differentiation. We used a 1.5-fold-change difference in expression between castes as the null model and genes were scored as significantly caste biased if they passed a FDR of 0.01 (i.e. 1.5 not overlapping with the 99% C.I.) in two-sided Wald tests.
Constructing cross-species co-expression network
A cross-species co-expression network was constructed using all caste-specific transcriptomes for the 6672-orthologous genes, normalized for colony-level variation. Co-expression level for each gene pair was then calculated based on the absolute values of Spearman’s correlation coefficients among all samples for gynes and workers, and the use of absolute values ensured that co-expression levels captured both positive regulation and negative regulation.
Projection of caste-specific single species transcriptomes on the principle component axes obtained for the five ant species with normal caste differentiation
After completing our analyses of caste-biased gene expression in the brains of the five ant species with normal queen-worker caste differentiation, we examined whether and to what extent the ant caste GRN might be shared with other social insect species by projecting brain published gene expression data of two queenless ants 22,27, the honeybee 41, and the paper wasp Polistes canadensis 27 onto the PCA axes obtained for our five focal ant species.
To validate this method, we reran the PCAs for different combinations of four of the five ants to test whether the PC axes produced would be consistent enough to also separate caste-biased gene expression in the fifth ant species. We did so by extracting the first two eigenvectors of the (quantile normalized and adjusted for species identity) transcriptomes of the four reference ant species using singular-value decomposition (SVD), which produced the first two PCs that separated castes across the four reference species. We then projected the normalized transcriptomes of the fifth ant species onto the PCA by matrix multiplication with the extracted eigenvectors from the four reference species. This procedure is comparable to using a leave-one-out jackknife resampling approach 58. Because it produced satisfactory results, we applied this procedure throughout comparisons between the five ant species with normal caste differentiation and the two queenless ants, the honey bee, and the paper wasp P. canadensis.
Supplementary Material
Acknowledgements
This work was supported by grants from the Lundbeck Foundation (GZ)(R190-2014-2827), the Carlsberg Foundation (G.Z.)(CF16-0663), the Strategic Priority Research Program of the Chinese Academy of Sciences (G.Z.)(XDB13000000), Biodiversity Research Center, Academia Sinica (J.W.) (100-2311-B-001-015-MY3, 103-2311-B-001-018-MY3, 104-2314-B-001-009-MY5), an Academia Sinica Career Development Grant (J.W.), and an ERC Advanced Grant (J.J.B.) (323085). We would like to thank Chunxue Guo, Hao Yu and Qiye Li for coordination of the sequencing at BGI.
Footnotes
Data availability
RNA-seq data have been deposited under Bioproject accession number PRJNA427677.
Contributions
G.Z., J.J.B. J.W. and B.Q. designed the experiments; R.S.L., N.-C.C., and B.Q. reared and isolated the ant colonies in the lab; B.Q. and N.-C.C. collected the ants, dissected the ant brains, and extracted RNA; B.Q. constructed the cDNA libraries; N.-C.C. and J.W. generated the transcriptome data for S. invicta; B.Q. analysed the data; B.Q. G.Z. and J.J.B. interpreted the data and wrote and revised the manuscript.
Competing interests
The authors declare no competing interests.
References
- 1.Wheeler WM. The ant-colony as an organism. Journal of Morphology. 1911;22:307–325. [Google Scholar]
- 2.Fisher RM, Cornwallis CK, West SA. Group Formation, Relatedness, and the Evolution of Multicellularity. Current Biology. 2013;23:1120–1125. doi: 10.1016/j.cub.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Boomsma JJ, Gawne R. Superorganismality and caste differentiation as points of no return: how the major evolutionary transitions were lost in translation. Biological Reviews. 2017;471:E1–54. doi: 10.1111/brv.12330. [DOI] [PubMed] [Google Scholar]
- 4.Ward PS. Ants. Current Biology. 2006 doi: 10.1016/j.cub.2006.02.054. [DOI] [PubMed] [Google Scholar]
- 5.Gould SJ. Ontogeny and Phylogeny. Harvard University Press; 1977. [Google Scholar]
- 6.Wagner GP. Homology, Genes, and Evolutionary Innovation. Princeton University Press; 2014. [DOI] [Google Scholar]
- 7.Simola DF, et al. Social insect genomes exhibit dramatic evolution in gene composition and regulation while preserving regulatory features linked to sociality. Genome Res. 2013;23:1235–1247. doi: 10.1101/gr.155408.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morandin C, et al. Comparative transcriptomics reveals the conserved building blocks involved in parallel evolution of diverse phenotypic traits in ants. Genome Biol. 2016;17:1. doi: 10.1186/s13059-016-1048-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toth AL, Robinson GE. Evo-devo and the evolution of social behavior. Trends Genet. 2007;23:334–341. doi: 10.1016/j.tig.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Carroll SB. Evo-Devo and an Expanding Evolutionary Synthesis: A Genetic Theory of Morphological Evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 11.Arendt D. The evolution of cell types in animals: emerging principles from molecular studies. Nat Rev Genet. 2008 doi: 10.1038/nrg2416. [DOI] [PubMed] [Google Scholar]
- 12.Barchuk AR, et al. Molecular determinants of caste differentiation in the highly eusocial honeybee Apis mellifera. BMC Developmental Biology. 2007;7:70. doi: 10.1186/1471-213X-7-70. 2007 7:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson, Tsutsui ND. Taxonomically restricted genes are associated with the evolution of sociality in the honey bee. BMC Genomics. 2011;12:164. doi: 10.1186/1471-2164-12-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feldmeyer B, Elsner D, Foitzik S. Gene expression patterns associated with caste and reproductive status in ants: worker-specific genes are more derived than queen-specific ones. Mol Ecol. 2014;23:151–161. doi: 10.1111/mec.12490. [DOI] [PubMed] [Google Scholar]
- 15.Sumner S. The importance of genomic novelty in social evolution. Mol Ecol. 2014;23:26–28. doi: 10.1111/mec.12580. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, Linksvayer TA. Deconstructing the Superorganism: Social Physiology, Groundplans, and Sociogenomics. Q Rev Biol. 2010;85:57–79. doi: 10.1086/650290. [DOI] [PubMed] [Google Scholar]
- 17.Ding Y, Zhou Q, Wang W. Origins of New Genes and Evolution of Their Novel Functions. Annu Rev Ecol Evol Syst. 2012;43:345–363. [Google Scholar]
- 18.Chen S, Krinsky BH, Long M. New genes as drivers of phenotypic evolution. Nat Rev Genet. 2013;14:645–660. doi: 10.1038/nrg3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warner MR, Mikheyev AS, Linksvayer TA. Genomic Signature of Kin Selection in an Ant with Obligately Sterile Workers. Mol Biol Evol. 2016;34:1780–1787. doi: 10.1093/molbev/msx123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ward PS. The phylogeny and evolution of ants. Annu Rev Ecol Evol Syst. 2014;24:2047–2052. [Google Scholar]
- 21.Mank JE. The transcriptional architecture of phenotypic dimorphism. Nature Ecology & Evolution. 2017;1:0006. doi: 10.1038/s41559-016-0006. [DOI] [PubMed] [Google Scholar]
- 22.Libbrecht R, Oxley PR, Keller L, Kronauer DJC. Robust DNA Methylation in the Clonal Raider Ant Brain. Current Biology. 2016:391–395. doi: 10.1016/j.cub.2015.12.040. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagner GP. The developmental genetics of homology. Nat Rev Genet. 2007;8:473–479. doi: 10.1038/nrg2099. [DOI] [PubMed] [Google Scholar]
- 24.Brawand D, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- 25.Roux J, Rosikiewicz M, Robinson-Rechavi M. What to compare and how: Comparative transcriptomics for Evo-Devo. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution. 2015;324:372–382. doi: 10.1002/jez.b.22618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lucas ER, Romiguier J, Keller L. Gene expression is more strongly influenced by age than caste in the ant Lasius niger. Mol Ecol. 2017;25:3389–5073. doi: 10.1111/mec.14256. [DOI] [PubMed] [Google Scholar]
- 27.Patalano S, et al. Molecular signatures of plastic phenotypes in two eusocial insect species with simple societies. PNAS. 2015;112 doi: 10.1073/pnas.1515937112. 201515937–13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monnin T, Ratnieks F, Jones GR, Beard R. Pretender punishment induced by chemical signalling in a queenless ant. Nature. 2002;419:61–65. doi: 10.1038/nature00932. [DOI] [PubMed] [Google Scholar]
- 29.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frumhoff PC, Ward PS. Individual-Level Selection, Colony-Level Selection, and the Association between Polygyny and Worker Monomorphism in Ants. ASN. 1992;139:559–590. [Google Scholar]
- 31.Schwander T, Rosset H, Chapuisat M. Division of labour and worker size polymorphism in ant colonies: the impact of social and genetic factors. Behav Ecol Sociobiol. 2005;59:215–221. [Google Scholar]
- 32.Trible W, Kronauer DJC. Caste development and evolution in ants: it's all about size. J Exp Biol. 2017;220:53–62. doi: 10.1242/jeb.145292. [DOI] [PubMed] [Google Scholar]
- 33.Nygaard S, et al. Reciprocal genomic evolution in the ant-fungus agricultural symbiosis. Nat Comms. 2016;7 doi: 10.1038/ncomms12233. 12233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monnin T, Peeters C. How many gamergates is an ant queen worth? Naturwissenschaften. 2007;95:109–116. doi: 10.1007/s00114-007-0297-0. [DOI] [PubMed] [Google Scholar]
- 35.Cronin AL, Molet M, Doums C, Monnin T, Peeters C. Recurrent Evolution of Dependent Colony Foundation Across Eusocial Insects. Annu Rev Entomol. 2013;58:37–55. doi: 10.1146/annurev-ento-120811-153643. [DOI] [PubMed] [Google Scholar]
- 36.Heinze J. The demise of the standard ant (Hymenoptera: Formicidae) Myrmecol News. 2008 [Google Scholar]
- 37.Rabeling C, Kronauer DJC. Thelytokous parthenogenesis in eusocial Hymenoptera. Annu Rev Entomol. 2013;58:273–292. doi: 10.1146/annurev-ento-120811-153710. [DOI] [PubMed] [Google Scholar]
- 38.Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics. 2009;10:161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peters RS, et al. Evolutionary History of the Hymenoptera. Current Biology. 2017;0:1013–1018. doi: 10.1016/j.cub.2017.01.027. [DOI] [PubMed] [Google Scholar]
- 40.Corona M, et al. Vitellogenin, juvenile hormone, insulin signaling, and queen honey bee longevity. PNAS. 2007;104:7128–7133. doi: 10.1073/pnas.0701909104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vleurinck C, Raub S, Sturgill D, Oliver B, Beye M. Linking Genes and Brain Development of Honeybee Workers: A Whole-Transcriptome Approach. PLoS ONE. 2016;11 doi: 10.1371/journal.pone.0157980. e0157980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.LaPolla JS, Dlussky GM, Perrichot V. Ants and the Fossil Record. Annu Rev Entomol. 2013;58:609–630. doi: 10.1146/annurev-ento-120710-100600. [DOI] [PubMed] [Google Scholar]
- 43.Barden P, Grimaldi DA. Adaptive Radiation in Socially Advanced Stem-Group Ants from the Cretaceous. Current Biology. 2016;26:515–521. doi: 10.1016/j.cub.2015.12.060. [DOI] [PubMed] [Google Scholar]
- 44.Peeters C. In: The Evolution of Social Behaviour in Insects and Arachnids. Crespi BJ, Choe JC, editors. Cambridge University Press; 1997. pp. 372–391. [Google Scholar]
- 45.Girardie J, Boureme D, Couillaud F, Tamarelle M, Girardie A. Anti-juvenile effect of neuroparsin A, a neuroprotein isolated from locust corpora cardiaca. Insect Biochemistry. 1987;17:977–983. [Google Scholar]
- 46.Toth AL, et al. Brain transcriptomic analysis in paper wasps identifies genes associated with behaviour across social insect lineages. Proc R Soc B. 2010;277:2139–2148. doi: 10.1098/rspb.2010.0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mikheyev AS, Linksvayer TA, Khaitovich P. Genes associated with ant social behavior show distinct transcriptional and evolutionary patterns. eLife Sciences. 2015;4 doi: 10.7554/eLife.04775. e04775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pantalacci S, et al. Transcriptomic signatures shaped by cell proportions shed light on comparative developmental biology. Genome Biol. 2017;18:29. doi: 10.1186/s13059-017-1157-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Romiguier J, et al. Phylogenomics Controlling for Base Compositional Bias Reveals a Single Origin of Eusociality in Corbiculate Bees. Mol Biol Evol. 2016;33:670–678. doi: 10.1093/molbev/msv258. [DOI] [PubMed] [Google Scholar]
- 50.Pontieri L, Schmidt AM, Singh R, Pedersen JS, Linksvayer TA. Artificial selection on ant female caste ratio uncovers a link between female-biased sex ratios and infection by Wolbachia endosymbionts. J Evol Biol. 2017;30:225–234. doi: 10.1111/jeb.13012. [DOI] [PubMed] [Google Scholar]
- 51.Conesa A, et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17:1. doi: 10.1186/s13059-016-1047-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keilwagen J, et al. Using intron position conservation for homology-based gene prediction. Nucl Acids Res. 2016;44:e89–e89. doi: 10.1093/nar/gkw092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lechner M, et al. Proteinortho: Detection of (Co-)orthologs in large-scale analysis. BMC Bioinformatics. 2011;12 doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nature Methods. 2017;14:417. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bourgon R, Gentleman R, Huber W. Independent filtering increases detection power for high-throughput experiments. PNAS. 2010;107:9546–9551. doi: 10.1073/pnas.0914005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bolstad BM, Irizarry RA, Åstrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003 doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 57.Leek JT. svaseq: removing batch effects and other unwanted noise from sequencing data. Nucl Acids Res. 2014;42:e161–e161. doi: 10.1093/nar/gku864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Efron B. Breakthroughs in Statistics. Springer; New York: 1992. pp. 569–593. [DOI] [Google Scholar]
- 59.Moreau CS, Bell CD, Vila R, Archibald SB, Pierce NE. Phylogeny of the Ants: Diversification in the Age of Angiosperms. Science. 2006;312:101–104. doi: 10.1126/science.1124891. [DOI] [PubMed] [Google Scholar]
- 60.Ward PS, Brady SG, Fisher BL, Schultz TR. The evolution of myrmicine ants: phylogeny and biogeography of a hyperdiverse ant clade (Hymenoptera: Formicidae) Systematic Entomology. 2015;40:61–81. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.