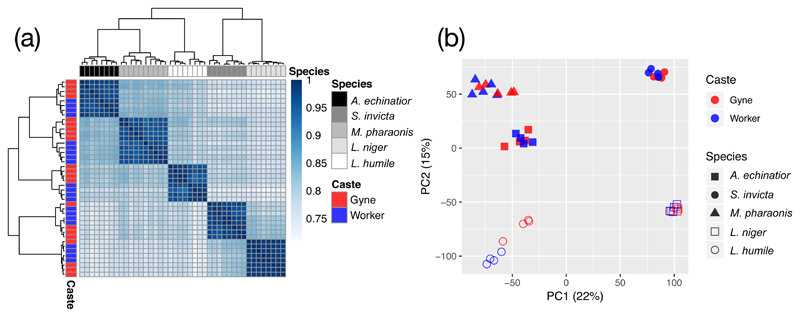
Figure 2. Gene expression signatures of species identity and caste type, calculated from 6672 one-to-one orthologs across seven ant species.
(a) Expression similarity matrix for brain samples with each cell representing overall transcriptomic expression similarity between a pair of samples based on Spearman correlation coefficients across all orthologous genes, ranging from 0 (white; no correlation) to 1 (dark blue; perfect positive correlation). The type of caste represented by each sample is illustrated in the red/blue left vertical bar and species identities are given in the grey-scale horizontal bar above the plot. (b) The first two principal components explaining transcriptomic variation across brain samples with red/blue colours for castes and different symbols for species: closed symbols refer to the three species of the sub-family Myrmicinae and open symbols to the remaining two species belonging to the sub-family Formicinae (L. niger) and Dolichoderinae (L. humile). All 6672 one-to-one orthologs were used for both plots, and expression levels were normalized by log2 transformation of the number of Transcripts per Million kilobases (TPM) and then by quantile normalization across samples.