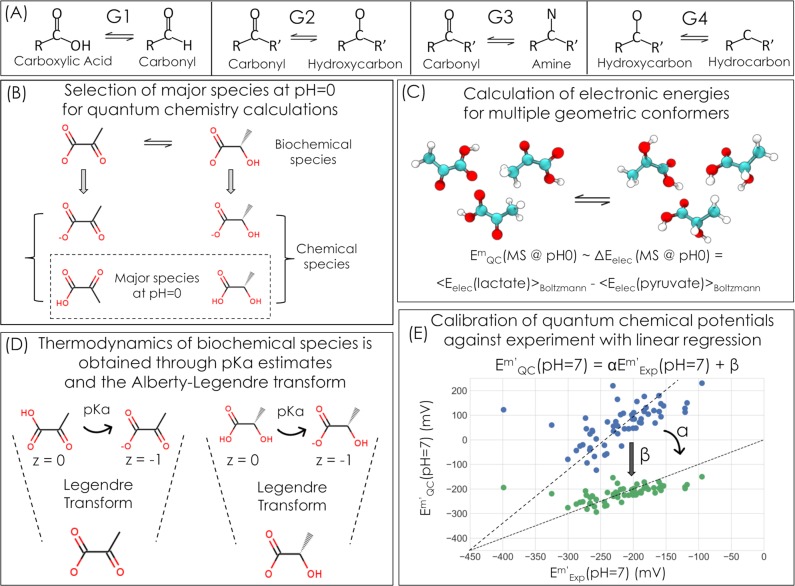
Fig 1. Our study is based on predicting biochemical standard redox potentials using a calibrated quantum chemistry strategy.
(A) The four different redox reaction categories considered here are reduction of a carboxylic acid to a carbonyl—G1, reduction of a carbonyl to a hydroxycarbon—G2, or an amine—G3, and reduction of a hydroxycarbon to a hydrocarbon—G4. (B) For each redox reaction of interest, such as reduction of pyruvate to lactate, we select the most abundant protonation state at acidic pH (pH = 0) for quantum chemical simulation. (C) We estimate the chemical redox potential as the difference between Boltzmann-averaged electronic energies of geometric conformers of products and substrates. (D) In order to convert chemical redox potentials to biochemical potentials at pH = 7, we use cheminformatic pKa estimates and the Alberty-Legendre Transform (Supplementary Information). (E) Finally, we use a set of 105 experimental values obtained from the NIST Thermodynamics of Enzyme-Catalyzed Reactions database (TECRDB) [30] and a set of Gibbs formation energies compiled by Robert Alberty [31] (Supplementary Information) to calibrate redox potentials using linear regression.