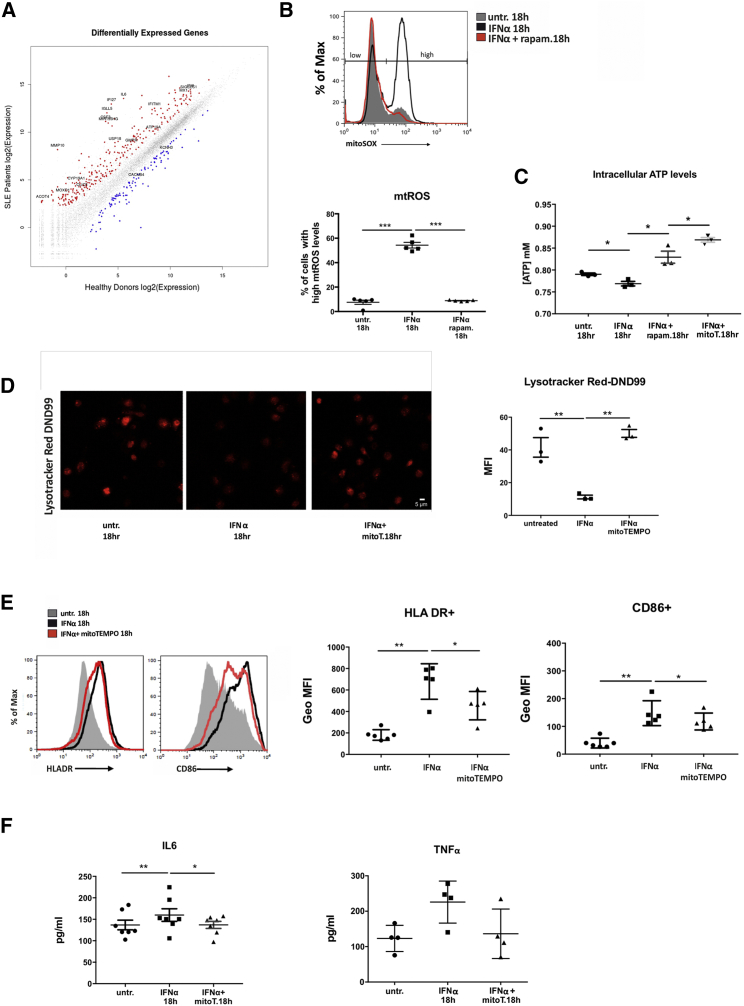
Figure 5.
Type I IFN Signaling Affects Mitochondrial Metabolism Leading to Proinflammatory Oxidative Stress and Ionic Alterations of Lysosomal Compartments
(Α) Scatterplot (R/Bioconductor) of log2-transformed RNA-seq expression values of all human genes between active SLE (n = 7) and healthy control (n = 5) CD14+ monocytes. All non-differentially expressed genes are depicted as gray colored. Selected genes related to type I IFN signature, oxidative stress, metabolism, antigen presentation, and differentiation are noted in the up (red color) and down (blue color) differentially expressed genes.
(B) Relative mitochondrial O2− levels of healthy CD14+ monocytes ± IFNα, rapam. were assessed by flow cytometry using mitoSOX dye (n = 5) One representative histogram overlay is shown and gates indicating low and high mtROS levels are depicted. Averages of % of cells in the mitoSOX high population are graphed. Datasets were analyzed using paired Student’s t test.
(C) Concentration of intracellular ATP concentration ± IFNα, rapam., or mitoTEMPO in lysates of CD14+ monocytes (n = 5). Datasets were analyzed using paired Student’s t test.
(D) Confocal microscopy for Lysotracker DND99 in CD14+ monocytes from healthy donors ± IFNα, mitoTEMPO. One representative result is depicted. Averages of MFI per cell are graphed (n = 3). Datasets were analyzed using paired Student’s t test.
(E) Levels of HLA-DR and CD86 expression measured by flow cytometry in CD14+ monocytes upon IFNα signaling and mtROS scavenging. GeoMFI averages are plotted (n = 3). Datasets were analyzed using paired Student’s t test.
(F) Concentrations of secreted IL6 and TNFα measured by ELISA in culture supernatants of CD14+ monocytes (n = 7) cultured as in (D) are graphed. Scale bar, 5 μM. Results are expressed as mean +SEM. ∗p < 0.05, ∗∗p < 0.005.