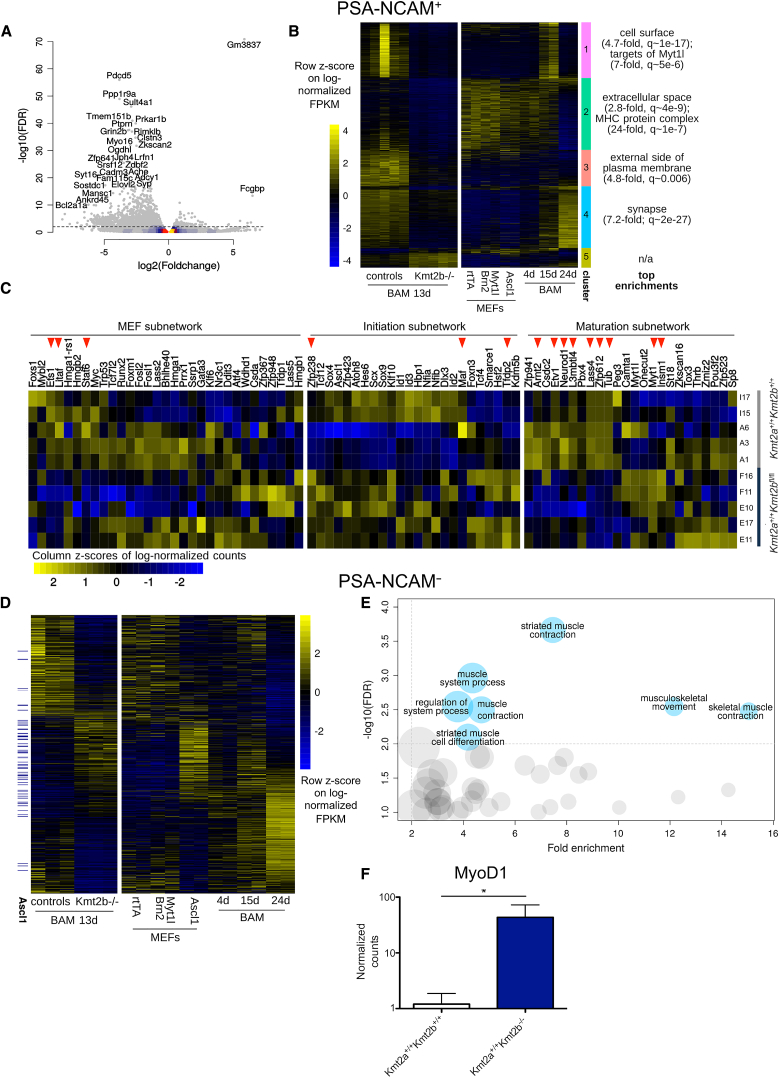
Figure 3.
KMT2B Loss Impairs Transcriptomic Changes Underlying Transdifferentiation
(A) Volcano plot highlighting the top DEGs among Kmt2b−/− and control 13-day iNs.
(B) DEGs upon Kmt2b KO clustered on the basis of their expression pattern across stages of transdifferentiation (plotted are log-normalized fragments per kilobase pair per million reads sequenced [FPKM], scaled across the whole dataset).
(C) Heatmap showing the expression across genotypes in iNs sorted at 13 days of the transcriptional regulators belonging to the three subnetworks described by Treutlein et al. (2016). The significant DEGs between Kmt2b−/− and the control are indicated by red arrows. Each line represents a different embryo from which the MEFs were derived.
(D) Expression pattern during normal transdifferentiation (right) and across genotypes on day 13 (left) of the DEGs found in 13-day Kmt2b−/− PSA-NCAM− cells.
(E) GO enrichment analysis of genes upregulated in 13-day Kmt2b−/− PSA-NCAM− cells with respect to controls.
(F) MyoD1 normalized counts in control and Kmt2b−/− PSA-NCAM− cells. Means ± SD are reported. ∗, FDR < 0.01.