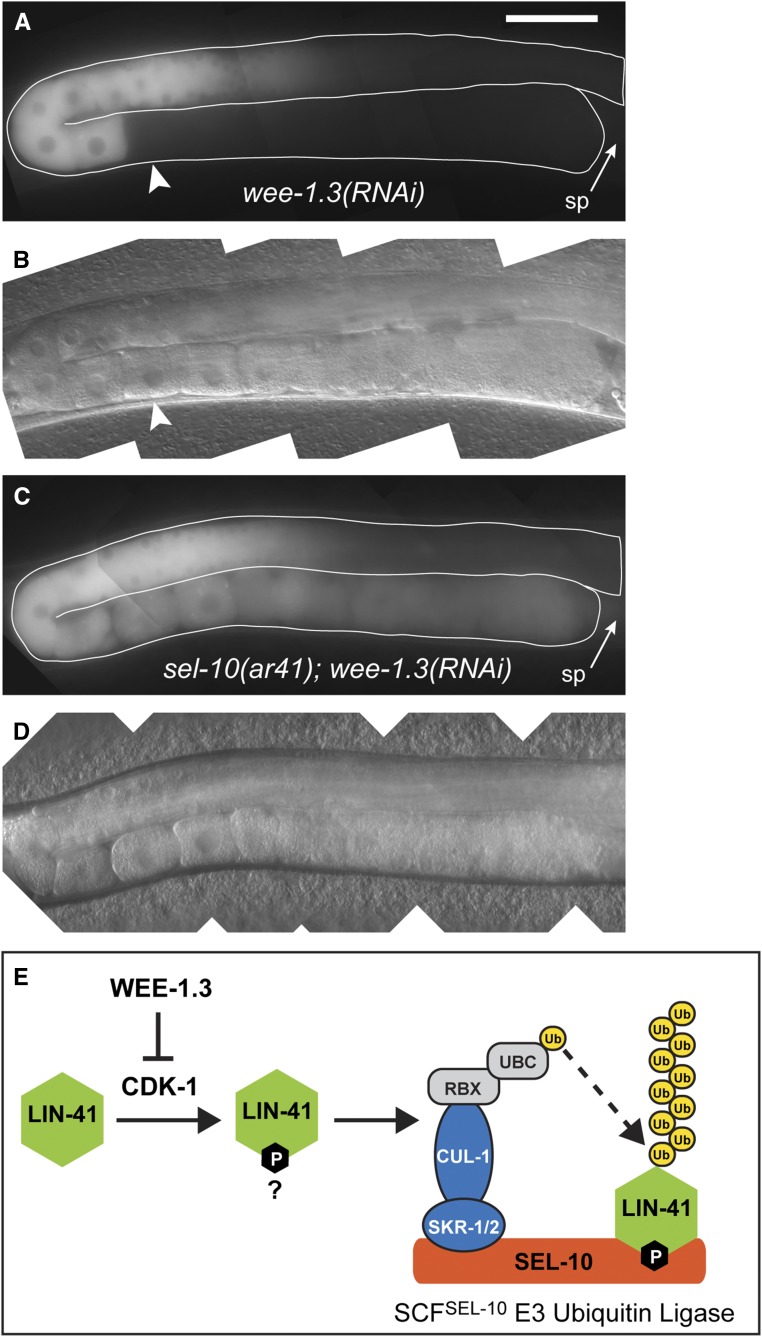
Figure 5.
SEL-10 is required for the WEE-1.3–inhibited degradation of GFP::LIN-41. (A–D) Composite GFP (A and C) and DIC (B and D) images of lin-41(tn1541); lon-3(e2175); wee-1.3(RNAi) (A and B) and lin-41(tn1541); lon-3(e2175) sel-10(ar41); wee-1.3(RNAi) (C and D) animals. GFP::LIN-41 is prematurely eliminated from oocytes by wee-1.3(RNAi) (arrowhead), but persists in abnormal oocytes near the spermatheca (sp; arrow) in sel-10(ar41); wee-1.3(RNAi) animals (C and D), suggesting that SEL-10 is required for this process; 150 ms GFP exposures, brightened slightly; Bar, 50 μm. (E) A simple model for the elimination of LIN-41 (green) that incorporates the known molecular functions of WEE-1.3 kinase, cyclin-dependent kinase (CDK-1) and subunits of the SCFSEL-10 E3 ubiquitin ligase. In brief, we hypothesize that SEL-10 (orange) may recognize phosphorylated LIN-41 (green) and trigger its ubiquitin-mediated degradation in collaboration with the other SCF E3 ubiquitin ligase subunits, SKR-1/2 (blue), and CUL-1 (blue). CUL-1 orthologs bind RING finger proteins (RBX; gray), which recruit a ubiquitin-conjugating enzyme (UBC; gray) that catalyzes the transfer of ubiquitin (yellow) to protein substrates, such as LIN-41. Subsequent recruitment of polyubiquitinated substrates to the proteasome results in degradation (not shown). This model is consistent with the epistatic relationship between wee-1.3(RNAi) and sel-10(ar41) with respect to the elimination of GFP::LIN-41, but other models are also possible.