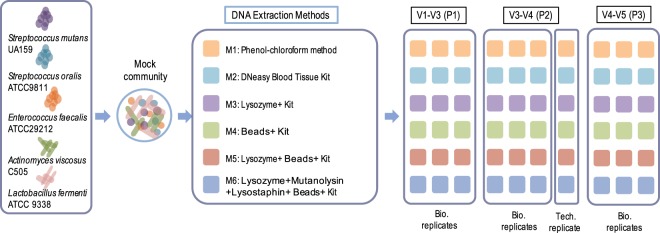
Figure 1.
Overall workflow of the study. Mock community samples consisting of five oral strains were sequenced via six DNA extraction methods and followed by amplification of three 16S rRNA gene target regions. The results of subsequent 16S rDNA sequencing on the MiSeq platform were quantitatively compared. For each of the six samples (each of which underwent one of the six DNA extraction method), triple biological replicates (“Bio. replicates”) for each of the amplification regions were sequenced. In addition, for the amplification region of V3-V4 (P2), one technical replicate (“Tech. replicate”) for each of the six samples was sequenced. All the 54 biological samples were processed by the same experimenter (Experimenter A) and sequenced on the same sequencing batch (Batch A), while six technical replicates were processed by the other experimenter (Experimenter B) and on the other batch (Batch A).