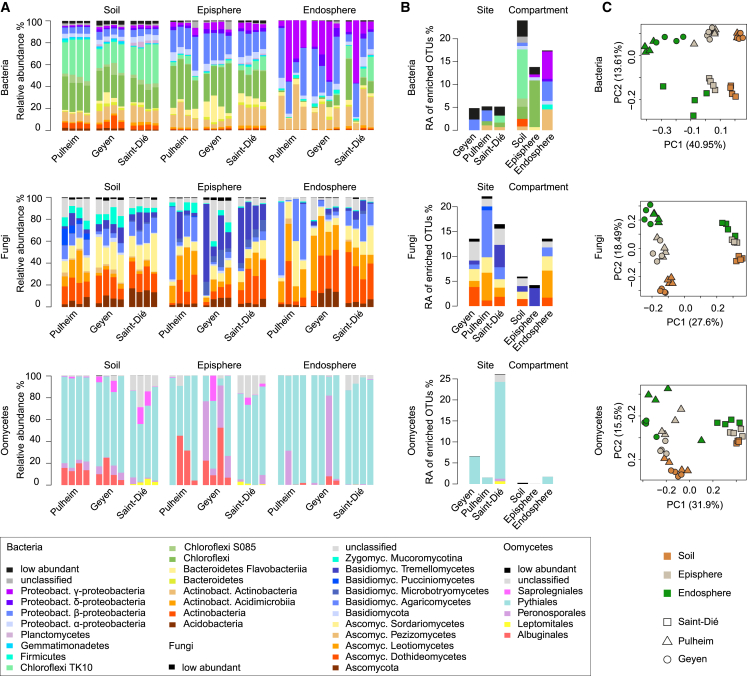
Figure 1.
Microbial Community Structure in Three Natural A. thaliana Populations
(A) Relative abundance (RA) of bacterial, fungal, and oomycetal taxa in soil, root episphere, and root endosphere compartments in three sites (Pulheim and Geyen in Germany and Saint-Dié in France). The taxonomic assignment is based on the Ribosomal Database Project (RDP) using a bootstrap cutoff of 0.5. Low-abundance taxonomic groups with less than 0.5% of total reads across all samples are highlighted in black. Each technical replicate comprised a pool of four plants.
(B) RA of OTUs significantly enriched in a specific site or compartment. A generalized linear model was used to compare OTU abundance profiles in one site or compartment versus the other two sites or compartments, respectively (p < 0.05, false discovery rate [FDR] corrected). The RAs for these OTUs were aggregated at the class level.
(C) Community structure of bacteria, fungi, and oomycetes in the 36 samples was determined using principal-component analysis. The first two dimensions of a principal-component analysis are plotted based on Bray-Curtis distances. Samples are color coded according to the compartment, and sites are depicted with different symbols.