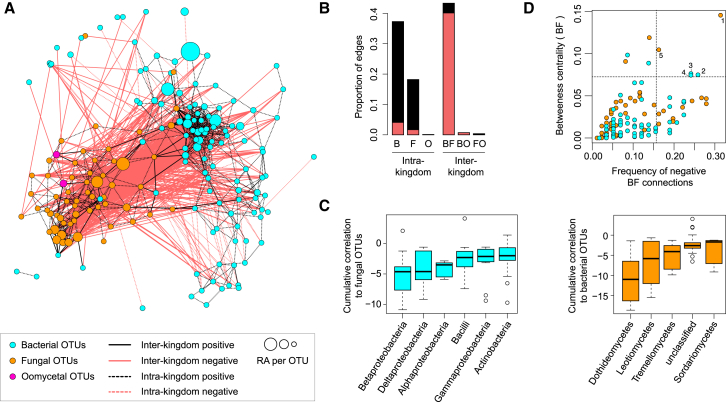
Figure 2.
Microbial Network of the A. thaliana Root Endosphere Microbiota
(A) Correlation-based network of root-associated microbial OTUs detected in three natural A. thaliana populations (Pulheim, Geyen, and Saint-Dié) in the endosphere. Each node corresponds to an OTU, and edges between nodes correspond to either positive (black) or negative (red) correlations inferred from OTU abundance profiles using the SparCC method (pseudo p < 0.05, correlation values < −0.6 or > 0.6). OTUs belonging to different microbial kingdoms have distinct color codes, and node size reflects their RA in the root endosphere compartment. Intrakingdom correlations are represented with dotted lines and interkingdom correlations by solid lines.
(B) Proportion of edges showing positive (black) or negative (red) correlations in the microbial root endosphere network. B, bacteria; F, fungi; O, oomycetes.
(C) Cumulative correlation scores measured in the microbial network between bacterial and fungal OTUs. Bacterial (left) and fungal (right) OTUs were grouped at the class level (>five OTUs per class) and sorted according to their cumulative correlation scores with fungal and bacterial OTUs, respectively.
(D) Hub properties of negatively correlated bacterial and fungal OTUs. For each fungal and bacterial OTU, the frequency of negative interkingdom connections is plotted against the betweenness centrality inferred from all negative BF connections (cases in which a node lies on the shortest path between all pairs of other nodes). The five microbial OTUs that show a high frequency of negative interkingdom connections and betweenness centrality scores represent hubs of the “antagonistic” network and are highlighted with numbers: (1) Davidiella, (2) Variovorax, (3) Kineosporia, (4) Acidovorax, and (5) Alternaria.