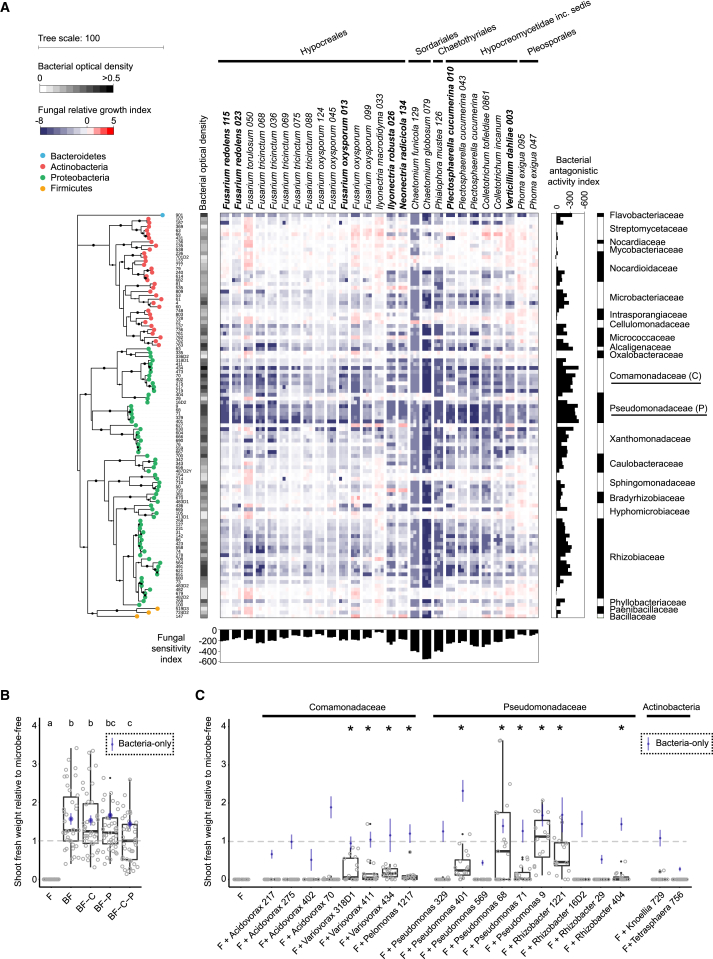
Figure 5.
Inhibitory Activities of Bacterial Root Microbiota Members toward Root-Associated Fungi
(A) Alteration of fungal growth upon interaction with phylogenetically diverse bacterial root commensals. The heatmap depicts the log2 fungal relativegrowth index (presence versus absence of bacterial competitors) measured by fluorescence using a chitin binding assay with wheat germ agglutinin (WGA), Alexa Fluor 488 conjugate. The phylogenetic tree was constructed based on the full bacterial 16S rRNA gene sequences, and bootstrapvalues aredepicted with black circles. Vertical and horizontal barplots indicate the cumulative antagonistic activity for each bacterial strain andthe cumulativesensitivity score for each fungal isolate, respectively. Alternating white and black colors are used to distinguish the bacterial families.All bacteria presented and 7/27 fungi (highlighted in bold) were used for the above-mentioned multi-kingdom reconstitution experiment (see Figure4).
(B) Shoot fresh weight of plants inoculated with fungi only (F) or bacteria plus fungi (BF, BF-C, BF-P, BF-C-P) relative to microbe-free control plants. -C, withdrawal of Comamonadaceae strains; -P, withdrawal of Pseudomonadaceae strains. Significant differences are depicted with letters (Kruskal-Wallis with Dunn’s post hoc test, p < 0.05). Relative shoot fresh weight of control plants inoculated with bacteria only is indicated for each strain (blue vertical lines; standard error, n = 8). Dashed gray horizontal line indicates shoot fresh weight of microbe-free plants normalized to 1.
(C) Same experiment as in (B), but instead of removing bacterial strains, individual bacterial isolates were co-inoculated with the 34-member fungal community (F) to determine their capacity for plant growth rescue.