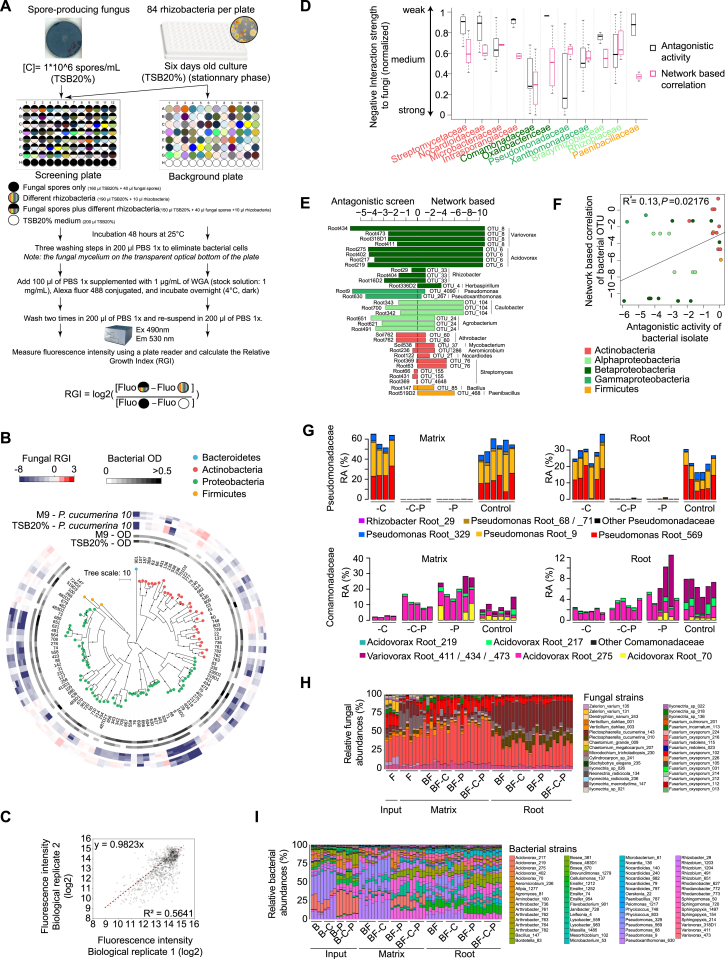
Figure S5.
High-Throughput Fungal-Bacterial Interaction Screen and Biocontrol Activities of Bacterial Root Commensals, Related to Figure 5
(A) Schematic overview of the experimental protocol. Fungal spores were equally distributed to the wells of a transparent-bottom 96 well plate and incubated in the presence or absence of different root-derived bacteria (stationary phase) in liquid medium (screening plate). Bacteria were also grown in the absence of fungal spores as a control (background plate). After 48 hours of interaction, three washing steps were used to eliminate bacterial cells in suspension. Note that the fungal mycelium sticks to the transparent optical bottom of the plate. After overnight incubation in Wheat Germ Agglutinin and two additional washes, the fluorescence intensity (reflecting fungal growth) was measured using a plate reader. The relative growth index was calculated as illustrated (see STAR Methods).
(B) Alteration of the growth of Plectosphaerella cucumerina isolate 10 upon competition with phylogenetically diverse members of the bacterial root microbiota in minimum medium (M9) and a carbon-rich medium (20% TSB). The phylogenetic tree was constructed based on the full bacterial 16S rRNA gene sequences and bootstrap values are depicted with black circles. The heatmap depicts the log2 fungal relative growth index (presence versus absence of bacterial competitors) measured by fluorescence (see above). Note the similar overall inhibitory activities in minimum and complex media.
(C) Validation experiment, in which the bacterial strains were re-screened against ten randomly-selected fungi. Fluorescence intensities (log2) were compared with those obtained from the first biological replicate.
(D) Comparison of network-derived correlations and experimentally tested interactions of bacterial families with fungal species. For each bacterial family shared between antagonistic screening and root-associated OTU network analysis, the average antagonistic activity against fungal isolates and the cumulative correlation to fungal OTUs in the network are represented. Bacterial families with more than two members were considered and values for both measurements were normalized to be in the same range.
(E) Direct comparison of bacterial OTUs from the root network with bacterial isolates used in the antagonistic screening. Each data point corresponds to a bacterial OTU-isolate pair (> 97% sequence similarity; only best matching hits are shown). For each pair, the network-derived correlation with fungal OTUs (from the bacterial OTU) is plotted against the result from the antagonistic screening (from the bacterial isolates).
(F) Same as E) but data are presented in a correlation plot.
(G) Validation of withdrawal of a subset of bacterial strains in the FlowPot microbiota reconstitution system. Relative abundances of isolates of the Pseudomonadaceae (top) and the Comamonadaceae (bottom) families in output matrix and root samples four weeks after inoculation (direct mapping at 100% sequence similarity). RA: Relative abundance. -C: withdrawal of ten Comamonadaceae strains, -P: withdrawal of eight Pseudomonadaceae strains. -C-P removal of 18 Comamonadaceae plus Pseudomonadaceae strains. Control samples were inoculated with the full 148-member bacterial community. Note that one Acidovorax strain (Acidovorax 275) was unintentionally not removed from the -C depleted samples. This Acidovorax strain is unable to rescue plant growth in the presence of the fungal community (see Figure 5C).
(H and I) Relative abundance of (H) fungal isolates and (I) bacterial isolates (direct mapping at 100% sequence similarity) is presented for all conditions. B: full 148-member bacterial community. F: 34-member fungal community. -C: depletion of Comamonadaceae isolates. -P: depletion of Pseudomonadaceae isolates. -C-P: depletion of Comamonadaceae and Pseudomonadaceae isolates.